Extraction and identification of membrane proteins from black widow spider eggs
Si-Ling FU, Jiang-Lin LI, Jia CHEN, Qiu-Ting WANG, Jian-Jun LI, Xian-Chun WANG,*
?
Extraction and identification of membrane proteins from black widow spider eggs
Si-Ling FU1, Jiang-Lin LI2, Jia CHEN1, Qiu-Ting WANG1, Jian-Jun LI1, Xian-Chun WANG1,*
1Key Laboratory of Protein Chemistry and Developmental Biology of Ministry of Education of China, Hunan Normal University, Changsha 410081, China2Molecular Sciences and Biomedicine Laboratory, State Key Laboratory for Chemo/Biosensing and Chemometrics, College of Biology, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha 410082, China
The eggs of oviparous animals are storehouses of maternal proteins required for embryonic development. Identification and molecular characterization of such proteins will provide much insight into the regulation of embryonic development. We previously analyzed soluble proteins in the eggs of the black widow spider (), and report here on the extraction and mass spectrometric identification of the egg membrane proteins. Comparison of different lysis solutions indicated that the highest extraction of the membrane proteins was achieved with 3%-4% sodium laurate in 40 mmol/L Tris-HCl buffer containing 4% CHAPS and 2% DTT (pH 7.4). SDS-PAGE combined with nLC-MS/MS identified 39 proteins with membrane-localization annotation, including those with structural, catalytic, and regulatory activities. Nearly half of the identified membrane proteins were metabolic enzymes involved in various cellular processes, particularly energy metabolism and biosynthesis, suggesting that relevant metabolic processes were active during the embryonic development of the eggs. Several identified cell membrane proteins were involved in the special structure formation and function of the egg cell membranes. The present proteomic analysis of the egg membrane proteins provides new insight into the molecular mechanisms of spider embryonic development.
;Egg;Membrane protein; Extraction; Identification
Introduction
In oviparous species, the egg is a storehouse of maternal proteins required for fertilization and the initiation of zygotic development (Calvert et al, 2003; Yue et al, 2011).During hatching, nutrients are transported from the egg components to the developing embryo. The proteins in the egg, which usually form lipoproteins with lipids, are crucial for the development, growth, and survival of the embryo (Laino et al, 2013; Wu et al, 2009; Zhao et al, 1994). Identification and molecular characterization of such proteins can provide insight into the regulation of embryonic development.Therefore, egg proteome analysis has become a hotspot in the field of proteomics. Up to now, many proteomic studies have been performed on eggs of the domestic chicken (Mann 2007; Rose & Hincke, 2009). Mann et al (2008) identified 119 proteins in egg yolk, 78 proteins in egg white, and 528 proteins in the decalcified eggshell organic matrix, whereas Farinazzo et al (2009) identified 255 yolk proteins in their study.
Cell membranes and cellular inner membranes are critical components of egg structure and function, and are involved in the partitioning of organelles, protecting the integrity of the genome and proteome, and providing defense against foreign molecules and external conditions that may damage or destroy the eggs. Membrane proteins are not only the main components of biological membranes but also the main executors of membrane functions. It has been estimated that membrane proteins account for about 30% of cellular proteins (Wu et al, 2003). Some proteomic analyses have focused on the membrane proteins of eggs. Due to the fact that most membrane proteins are hydrophobic and difficult to dissolve in aqueous buffer, the extraction of egg membrane proteins is often performed with the help of detergents and/or other additives (Mann et al, 2006; Rose & Hincke, 2009). For example, SDS-containing lysis solution was used to solubilize proteins in the hen eggshell membrane (ESM), followed by SDS-PAGE and LC-MS/MS, revealing 62 proteins, including 53 not previously reported (Kaweewong et al, 2013). Egg proteomes of other species, such as the silkworm (Fan et al, 2013) and snail (Sun et al, 2012), have also been analyzed. However, reports on spider egg proteomes are limited.
The black widow spider is an oviparous animal. The adult female constructs 7-8 egg sacs containing about 450 eggs each from June to October, with 1-3 weeks in between (Bonnet 2004). Compared with the eggs of other species, the eggs of the black widow spider received particularly early attention due to their inherent toxicity (Kobert 1889; Buffkin et al, 1971; Russell & Maret?, 1979). In our previous work, we conducted a proteomic analysis of the water-soluble proteins in the eggs of the black widow spider () and identified 157 proteins, concluding that the molecular basis and mechanism for egg toxicity were different from those of spider venom (Li et al, 2012). Our present work analyzed the membrane proteins in the spider eggs based on detergent-assisted extraction and mass spectrometric identification to more comprehensively understand the egg proteome of the spider.
Materials and methods
Reagents
Dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate, SDS, 3-[(3-cholamidopropyl) dimethylamino] propanesulfonate (CHAPS), acrylamide, bisacrylamide, glycine and Tris were purchased from Sigma-Aldrich (St. Louis, MO, USA).Sodium laurate (SL) was from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Trypsin was from Promega (Madison, WI, USA). Protein assay kit was from Solarbio (Beijing, China). All other reagents were products of the highest grade available.
Extraction of egg membrane proteins
The eggs were homogenized in ddH2O with a mortar and pestle and the resulting homogenate was centrifuged at 15 400 g for 15 min at 4 °C. After the supernatant was removed, the pellet (membrane debris) was repeatedly homogenized and extensively washed to eliminate the soluble components, and was then lyophilized. For extracting proteins from the egg membrane debris, different lysis solutions (40 mmol/L Tris-HCl/4% CHAPS/2% DTT, containing 1%-4% of SDS or SL, pH 7.4) were used and compared. Briefly, 50 mg aliquots of the lyophilized membrane debris were separately placed in eight Eppendorf tubes, with the eight different lysis solutions added, respectively. After intermittent oscillation with a vortex oscillator for 30 min, the mixtures were centrifuged at 15 400 g for 15 min. The supernatants were separately collected and the pellets were extracted twice. The obtained supernatants were separately pooled and the total volume of the extraction for each tube was adjusted to 2.5 mL. Protein content was determined with the protein assay kit using BSA as the standard according to the manufacturer’s instructions.
SDS-PAGE of egg membrane proteins
SDS-PAGE of the egg membrane proteins was performed according to Laemmli (1970) under denaturing conditions on a 10% polyacrylamide separating gel overlaid with a 5% stacking gel. The sample solution (6 μL) was mixed with 3μL of loading sample buffer (500 mmol/L Tris-HCl, 4% SDS, 100 mmol/L DTT, 20% glycerol, a trace of bromophenol blue, pH 6.8,) and then centrifuged at 10000 gfor 10 min. The proteins in the supernatants were loaded and separated through gel electrophoresis, which was run at 20 mA on the staking gel and at 40 mA on the separating gel. After completion of electrophoresis, the resolved proteins in the gel were fixed with 10% acetic acid/40%methanol and visualized by staining with Coomassie brilliant blue G-250.
In-gel digestion
Digestion of the separated proteins in the gel was performed according to Chen et al (2006). Briefly, the lane gel was cut into slices about 2-3 mm wide and then broken into small pieces, followed by washing with 25 mmol/L NH4HCO3and 50% ACN/25 mmol/L NH4HCO3sequentially. The gel pieces were dehydrated in 100% ACN and then dried in a Speed Vac. In-gel digestion was performed with trypsin (1 μg enzyme/slice) in 25 mmol/L NH4HCO3containing 10% ACN with incubation overnight at 37 °C. The released peptides were extracted twice by adding 100 μL of 67% ACN containing 5% formic acid with ultrasonication. The supernatants were pooled and then concentrated in a Speed Vac.
nLC-MS/MS analysis
Tryptic digests prepared by in-gel digestion were analyzed by an-nLC system (Proxeon Biosystems, Odense, Denmark) coupled with a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA, USA) and HCTultra ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany), equipped with an autosampler and a C18 reverse phase column (PepMap, 75 μm i.d., 15 cm long, Sunnyvale, CA, USA). For nLC separation, solvent A (0.1% formic acid) and solvent B (0.1% formic acid in ACN) were used. The peptides were eluted using a gradient from 0 to 35% B in 38 min, 35% to 90% B in 15 min, and 90% to 100% B in 5 min. The flow rate was 200 nL/min. The peptides eluted from the column were online directed into the mass spectrometer. The LC-MS system was controlled by Data-Dependent Automatic Acquisition (DDA), using positive ion detection mode. Peptide ions were detected in the MS scan, and the seven most abundant ions in each MS scan were selected for collision-induced dissociation (CID) using data-dependent MS/MS mode over the m/z range of 350-1800.
Data processing and protein identification
Raw mass spectrometry data were processed with Xcalibur v.2.1 (Thermo Scientific) and Proteome Discoverer v.1.3 beta (Thermo Scientific). For protein identification, database searches were performed using the in-house sequence algorithm in the Proteome Discoverer software against UniProt/Swiss-Prot and UniProt/TrEMBL, with the parameters set as follows: enzyme, trypsin; allowance of up to two missed cleavages; MS accuracy, 0.015; MS/MS accuracy, 0.05; fixed modification, carbamidomethylation (C); variable modification, oxidation (Met).Proteins were identified at the 95% confidence level (<0.05). The theoretical molecular weight (MW) and isoelectric point (pI) of the identified proteins were retrieved from the output files of Proteome Discoverer. Further information on the subcellular location and function of identified proteins were retrieved from http://www.uniprot.org/uniprot.
Results
Extraction and separation of egg membrane proteins
Due to the water-insolubility of the membrane proteins in black widow spider eggs, we used eight mixed lysis solutions containing different concentrations (1%-4%) of SL or SDS to extract this set of proteins. The protein extraction efficiencies were compared and demonstrated different extraction abilities, as shown in Table 1. As the concentration of the detergent increased, the protein extraction rate gradually increased. SL-containing lysis solutions achieved extraction rates varying from 1.28% to 2.33%, whereas the protein extraction rate of SDS-containing lysis solutions was 1.33% to 1.97%. Figure 1 shows the dynamic changes in extraction efficiency based on the protein concentration of the extract. Comparatively, when the concentrations of SDS and SL were 1% and 2%, the protein extraction efficiencies of the two lysis solutions were not significantly different (>0.05). However, when the concentrations of the detergents increased to 3% and 4%, the extraction efficiency of the SL-containing mixed lysis solutions was higher than that of the SD-containing mixed lysis solutions (<0.05). The highest extraction of egg membrane proteins was obtained with the mixed lysis solution containing 3%-4% SL.
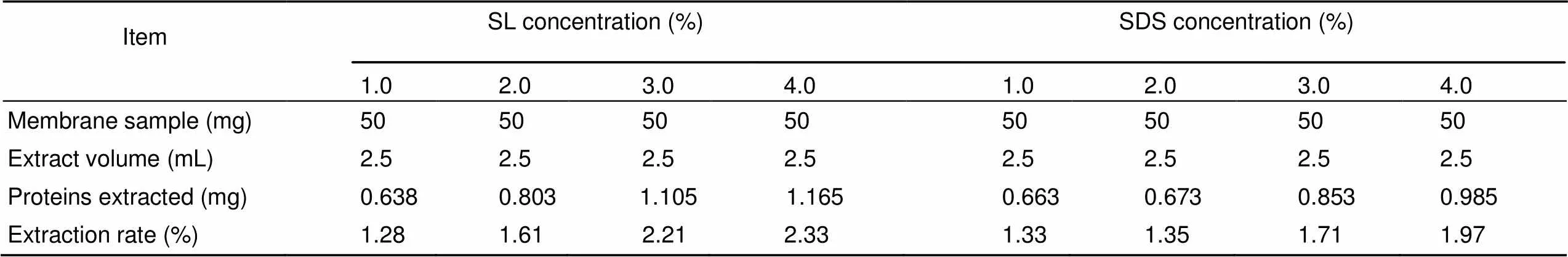
Table 1 Comparison of the extraction efficiencies of different mixed lysis solutions
Figure 1 Effect of different mixed lysis solutions on the extraction of egg membrane proteins
The experiments were performed in triplicate.
Prior to nLC-MS/MS analysis, the extracted membrane protein mixtures were separated in parallel lanes with SDS-PAGE (Figure 2), which showed that the protein distribution profiles were similar between the two extraction conditions. As the concentration of the detergents increased, the protein content of the extract exhibited an increasing trend. However, when the concentrations of the detergents were greater than 2%, the extraction of proteins greater than about 60×103was obviously enhanced while that of the proteins below 60×103remained relatively stable, indicating that the efficient extraction of higher molecular weight proteins, which are generally more hydrophobic, needed detergents at higher concentrations.
Figure 2 SDS-PAGE image of egg membrane proteins extracted by different mixed lysis solutions
Lanes 1-4: proteins extracted by lysis solutions containing 1%, 2%, 3%, and 4% SDS, respectively; Lanes 5-8: proteins extracted by lysis solutions containing 1%, 2%, 3%, and 4% SL, respectively; Lane 9: protein molecular weight marker.
Identification the membrane proteins
After the proteins in the gel slices were in-gel digested and the recovered tryptic peptides were analyzed with nLC-MS/MS, the acquired data were used to search against protein databases. As a result, a total of 39 proteins with membrane-localization information were identified after de-redundancy and removal of the proteins with no membrane localization information (Table 2). The data inTable 2 and Figure 3 show that the identified membrane proteins were distributed in the MW range of 14.6×103-352.9×103, with more than 60% of the identified membrane proteins in the MW range of 20×103-80×103. The 39 membrane proteins were distributed in the pI range of 4.74-10.04. About 77% of the proteins were acidic (pI<7), with proteins with a pI value between 5 and 6 accounting for 53.85% (Figure 4). Table 2 also lists the number of unique peptides identified for each protein. The proteins keratin, hornerin, and ATP synthase had relatively higher numbers of unique peptides.

Table 2 Information on the membrane proteins identified from the spider eggs
No. of pep: Number of unique peptide for an identified protein.
According to their biological functions, the 39 identified membrane proteins could be divided into four groups, though this functional classification was not strict as proteins usually have multiple functions: (i) Metabolic enzyme; (ii) Structure; (iii) Regulation; and (iv) Transport (Figure 5; Table 2). Nineteen membrane proteins (accounting for 48.72%) were metabolic enzymes, involved in substance and energy metabolism, including acetyl-coenzyme A carboxylase carboxyl transferase, ATP synthase and carbamoyl-phosphate synthase. Eleven proteins (28.21%) were classified into the “Structure” group, and included membrane-localized cytoskeleton components and cell membrane proteins related to cell adhesion and cell junction, such as hornerin, plakophilin-1, junction plakoglobin, and some special kinds of keratins. Most proteins in this group not only acted as a structural component but also exerted otherbiological functions (see Discussion). Six regulatory proteins and three transport proteins were unambiguously identified, and were primarily involved in the regulation of enzyme activity and signal pathways as well as transmembrane transport of proteins.
Figure 3 Distribution of the identified membrane proteins as a function of molecular weight (MW)
Values on the top of the column are the number of identified proteins.
Figure 4 Distribution of identified membrane proteins as a function of isoelectric point (pI)
Values on the top of the column are the number of identified proteins.
Figure 5 Functional classification of the identified membrane proteins
Of the 39 identified membrane proteins, 14 had unambiguous cell membrane-localized annotation in the protein databases. These cell membrane proteins were involved in catalytic, structural and regulatory functions. At least eight identified cell membrane proteins, including hornerin, plakophilin-1, junction plakoglobin and neuronal growth regulator 1, were involved in membrane cornification and recognition and communication between eggs, suggesting that the cell membrane of these spider eggs had specificities.
Discussion
Contemporary proteomic techniques provide an effective means to comprehensively analyze all proteins in a cell, tissue or organism. However, membrane protein analysis has comparatively lagged behind that of soluble proteins due to their poor solubility in aqueous buffers, which limits their solubilization, extraction and enzymolysis (Rabilloud 2003; Santoni et al, 2000; Wu & Yates, 2003). To improve the extraction of membrane proteins, particularly high-hydrophobicity integral membrane proteins, a series of additives can be supplemented to the buffer alone or in combination, including detergents, chaotropes, aqueous-organic solvents, and organic acids (Masuda et al, 2008; Speers & Wu, 2007). SDS is the most often used detergent because it has a stronger ability than other commonly used detergents to disrupt biological membranes and extract membrane proteins (Masuda et al, 2008; Botelho et al, 2010). However, SDS is difficult to remove from the samples and therefore interferes with the following proteolysis and mass spectrometric analysis, thus limiting its application to membrane proteomics (Yu et al, 2003; Botelho et al, 2010). Recently, another detergent, sodium laurate (SL), was found not only to lyse membranes and extract membrane proteins as efficiently as SDS, but was also compatible with proteases and mass spectrometry as it can be conveniently removed from samples by phase transfer after acidification (Lin et al, 2013). In the present study, we used SDS- and SL-containing lysis solutions to extract the membrane proteins from black widow spider eggs. Because the eggs used were near hatching, their cell membranes were cornified to a certain extent, which further hampered the lysis of membranes and the extraction of membrane proteins. Therefore, to enhance extraction efficiency, other additives (CHAPS and DTT) were added to the Tris-HCl buffer to constitute a basic lysis solution. The results demonstrated that the mixed lysis solution containing higher concentrations of SL showed a certain advantage over the SDS-containing mixed lysis solution in the extraction of egg membrane proteins.
Membrane proteins perform a variety of cellular functions, including catalyzing metabolic reactions, transporting molecules and ions across the membrane, relaying signals in metabolic regulation and allowing cells to identify and interact with each other (Almén et al, 2009). In the present study, we identified 39 membrane proteins with corresponding functions. These proteins were distributed in wide MW (14.6×103-352.9×103) and pI (4.74-10.04) ranges, suggesting diversity in both structure and function. Nearly half of the identified membrane proteins were metabolic enzymes, the majority of which were involved in energy metabolism and synthetic metabolism and included ATP synthase, succinate dehydrogenase, adenylate kinase, nucleoside diphosphate kinase, acetyl-coenzyme A carboxylase carboxyl transferase, aspartate carbamoyltransferase, and adenylosuccinate synthetase. The identification of these metabolic enzymes indicated that the relevant metabolic processes were active during the embryonic development of the spider egg. All metabolic processes in a cell are elaborately regulated and many membrane proteins participate in these regulation processes.
Six identified proteins had regulatory activities and were classified into the “Regulation” group, although they also had other biological functions. For example, 14-3-3 protein epsilon in this group is a member of the 14-3-3 protein family and is involved in the regulation of a large spectrum of cellular processes, including metabolism, signal transduction, and cell development (Dunaway et al, 2005; Paul & van Heusden, 2005). Many experiments have demonstrated the importance of 14-3-3 proteins in embryonic development. Wu & Muslin (2002) used an unphosphorylated peptide inhibitor of 14-3-3, R18, to determine the role of 14-3-3 proteins inembryonic development, and demonstrated the requirement for 14-3-3 in mesodermal specification. Inhibition of 14-3-3 resulted in embryos with axial patterning defects and reduced expression of mesodermal marker genes. These phenotypic defects were caused by impaired fibroblast growth factor signaling in R18-injected embryos. To evaluate the role of individual 14-3-3 proteins in vertebrate embryonic development, Lau & Muslin (2009) utilized an antisense morpholino oligo microinjection technique inembryos, and showed that embryos lacking specific 14-3-3 proteins displayed unique phenotypic abnormalities. Keratin, type II cytoskeletal 1 was another identified membrane protein classified into this group, which not only acts as a structural constituent of the cell membrane but also as a high affinity receptor (Pixley et al, 2011).
Eleven membrane proteins were categorized into the “Structure” group, including several keratins closely related to embryo development. Lu et al (2005) demonstrated that two type I and two type II keratin genes were already transcribed in the 2-cell stage embryo, and type II keratins preceded type I keratins during early embryonic development. Hornerin was a constituent of the cell membrane and was identified based on 10 unique peptides, only lower than that of several keratins, suggesting that the concentration of hornerin in the sample was fairly high. The relative concentration of a protein identified by mass spectrometry is directly related to the number of identified peptides, neglecting the possible effects of other factors such as enzymatic digestion constraint, detection mass range of the mass spectrometer and differential post-translational modification. Therefore, the number of identified unique peptides assembled into a protein may reflect the protein’s relative abundance (Fan et al, 2013; Jin et al, 2008; Li et al, 2010). Hornerin can bind Ca2+and was rich in the cell membrane in our present study, suggesting that it may play an important role in the cornification of cell membrane because Ca2+is known to trigger the process of cornification (Hennings et al, 1980). Another identified cell membrane protein, desmocollin-1, can also bind Ca2+and is involved in cornification (Ishida-Yamamoto et al, 2011). During embryo development, calcium is a major nutritional requirement (Johnston & Comar, 1955). It is speculated that the existence of such Ca2+-binding proteins in the cell membrane not only enrolls Ca2+to strengthen the cell membrane, but also stores Ca2+for later stages of embryo development. It is worth mentioning that we identified several cell membrane proteins with cell adhesion and cell junction functions, including plakophilin-1, desmoglein-1 and junction plakoglobin, suggesting recognition and communication between eggs in the same egg sac, although the eggs existed dispersedly. In addition, the identification of polyribonucleotide nucleotidyltransferase and several proteins with protein transport activity, as well as chaperonin, suggested that processing and translation of mRNA was active during the embryonic development of the eggs. A batch of ribosomal proteins and regulatory factors for protein synthesis were identified, together with membrane proteins from the eggs, supporting this speculation (data not shown).
In summary, to efficiently extract and identify the membrane proteins of black widow spider eggs, we comparatively employed different lysis solutions to lyse the biological membranes and extract membrane proteins, followed by SDS-PAGE and nLC-MS/MS analysis. The mixed lysis solution containing SL showed a certain advantage over that containing SDS when the concentrations of the detergents were higher (3%-4%). A total of 39 membrane proteins involved in structure, catalysis, metabolism regulation, signal transduction or cell communication were identified, which is consistent with the functions of biological membranes. Nearly half of the identified membrane proteins were metabolic enzymes involved in various cellular processes, particularly energy metabolism and biosynthesis, suggesting that relevant metabolic processes were active during the embryonic development of the eggs. The identification of cell membrane proteins is helpful for revealing the special structure and functions of egg cell membranes.
REFERENCES
Almén MS, Nordstr?m KJ, Fredriksson R, Schi?th HB. 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin.7: 50-63.
Bonnet MS. 2004. The toxicology of: the mediterranean black widow spider.93(1): 27-33.
Botelho D, Wall MJ, Vieira DB, Fitzsimmons S, Liu F, Doucette A. 2010. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation.9(6): 2863-2870.
Buffkin DC, Russell FE, Deshmukh A. 1971. Preliminary studies on the toxicity of black widow spider eggs.9(4): 393-402.
Calvert ME, Digilio LC, Herr JC, Coonrod SA. 2003. Oolemmal proteomics – identification of highly abundant heat shock proteins and molecular chaperones in the mature mouse egg and their localization on the plasma membrane.1: 27-34.
Chen P, Li XW, Sun Y, Liu Z, Cao R, He QY, Wang MC, Xiong JX, Xie JY, Wang XC, Liang SP. 2006. Proteomic analysis of rat hippocampal plasma membrane: characterization of potential neuronal- specific plasma membrane proteins.98(4): 1126-1140.
Dunaway S, Liu HY, Walworth NC. 2005. Interaction of 14-3-3 protein with Chk1 affects localization and checkpoint function.118: 39-50.
Fan LF, Lin JR, Zhong YS, Liu YJ. 2013. Shotgun proteomic analysis on the diapause and non-diapause eggs of domesticated silkworm Bombyx mori.8(4): e60386.
Farinazzo?A, Restuccia U, Bachi A, Guerrier L, Fortis F, Boschetti E, Fasoli E, Citterio A, Righetti PG. 2009. Chicken egg yolk cytoplasmic proteome, mined via combinatorial peptide ligand libraries.1216(8): 1241-1252.
Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. 1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture.19(1): 245-254.
Ishida-Yamamoto A, Igawa S, Kishibe M. 2011. Order and disorder in corneocyte adhesion.38(7): 645-654.
Jin SS, Daly DS, Springer DL, Miller JH. 2008. The effects of shared peptides on protein quantitation in label-free proteomics by LC/MS/MS., 7(1): 164-169.
Johnston PM, Comar CL. 1955. Distribution and contribution of calcium from the albumen, yolk and shell to the developing chick embryo.183(3): 365-370.
Kaweewong K, Garnjanagoonchorn W, Jirapakkul W, Roytrakul S. 2013. Solubilization and identification of hen eggshell membrane proteins during different times of chicken embryo development using the proteomic approach.32(4): 297-308.
Kobert R. 1889. Ueber die giftigen Spinnen Russlads., 8: 340-362.
Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4.227(5259): 680-685.
Laino A, Cunningham M, Costa FG, Garcia CF. 2013. Energy sources from the eggs of the wolf spider: isolation and characterization of lipovitellins.165(3): 172-180.
Lau JMC, Muslin AJ. 2009. Analysis of 14-3-3 family member function inembryos by microinjection of antisense morpholino oligos.518: 31-41.
Li JJ, Liu H, Duan ZG, Cao R, Wang XC, Liang SP. 2012. Protein compositional analysis of the eggs of black widow spider (): implications for the understanding of egg toxicity.26(12): 510-515.
Li JY, Moghaddam SH, Chen JE, Chen M, Zhong BX. 2010. Shotgun proteomic analysis on the embryos of silkwormat the end of organogenesis.40(4): 293-302.
Lin Y, Huo LJ, Liu ZH, Li JL, Liu Y, He QZ, Wang XC, Liang SP. 2013. Sodium laurate, a novel protease- and mass spectrometry-compatible detergent for mass spectrometry-based membrane proteomics.8(3): e59779.
Lu H, Hesseb M, Peters B, Magina TM. 2005. Type II keratins precede type I keratins during early embryonic development.84(8): 709-718.
Mann K. 2007. The chicken egg white proteome.7(19): 3558-3568.
Mann K, Ma?ek B, Olsen JV. 2006. Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer.6(13): 3801- 3810.
Mann K, Olsen JV, Ma?ek B, Gnad F, Mann M. 2008. Identification of new chicken egg proteins by mass spectrometry-based proteomic analysis.64(2): 209-218.
Masuda T, Tomita M, Ishihama Y. 2008. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis.7(2): 731-740.
Paul G, van Heusden H. 2005. 14-3-3 proteins: regulators of numerous eukaryotic proteins.57(9): 623-629.
Pixley RA, Espinola RG, Ghebrehiwet B, Joseph K, Kao A, Bdeir K, Cines DB, Colman RW. 2011. Interaction of high-molecular-weight kininogen with endothelial cell binding proteins suPAR, gC1qR and cytokeratin 1 determined bysurface plasmon resonance (BiaCore).105(6): 1053-1059.
Rabilloud T. 2003. Membrane proteins ride shotgun.21(5): 508-510.
Rose MLH, Hincke MT. 2009. Protein constituents of the eggshell: eggshell-specific matrix proteins.66(16): 2707-2719.
Russell FE, Maret? Z. 1979. Effects ofegg poison on web building.17(6): 649-650.
Santoni V, Molloy M, Rabilloud T. 2000. Membrane proteins and proteomics: Un amour impossible?21(6): 1054-1070.
Speers AE, Wu CC. 2007. Proteomics of integral membrane proteins-theory and application.107(8): 3687-3714.
Sun J, Zhang HM, Wang H, Heras H, Dreon MS, Ituarte S, Ravasi T, Qian PY, Qiu JW. 2012. First proteome of the egg perivitelline fluid of a freshwater gastropod with aerial oviposition.11(8): 4240-4248.
Wu CC, Yates III JR 2003. The application of mass spectrometry to membrane proteomics.21(3): 262-267.
Wu CC, MacCoss MJ, Howell KE, Yates III JR. 2003. A method for the comprehensive proteomic analysis of membrane proteins.21(5): 532-538.
Wu CL, Muslin AJ. 2002. Role of 14-3-3 proteins in earlydevelopment.119(1): 45-54.
Wu XG, Liu ZJ, Yao GG, Cheng YX, Yang XZ, Wang CL. 2009. Relationship between the organogenesis of hepatopancreas and the yolk utilization during embryonic development of swimming crab,.30(4): 499-456. (in Chinese)
Yu YQ, Gilar M, Lee PJ, Bouvier ESP, Gebler JC. 2003. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins.75(21): 6023-6028.
Yue MJ, Mo SJ, Song P, Gong YZ. 2011. Identification of Z-OTU protein during zebrafish oogenesis and early embryogenesis.32(4): 386-390. (in Chinese)
Zhao XF, Wang JX, Li M. 1994. The study on the proteinases responsible for the degradation of yolk proteins in15(S1): 119-123. (in Chinese)
Received: 20 April 2015; Accepted: 08 June 2015
This work was supported by grants from the National Natural Science Foundation of China (31271135, 31070700), the National Innovation Experimental Program for University Students (201310542008) and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486)
, E-mail: wang_xianchun@263.net
10.13918/j.issn.2095-8137.2015.4.248
- Zoological Research的其它文章
- Effects of frugivorous birds on seed retention time and germination in Xishuangbanna, southwest China
- The breeding biology of Red-Whiskered Bulbul (Pycnonotus jocosus) in Xishuangbanna, southwest China
- Population genetic studies in the genomic sequencing era
- Why do we study animal toxins?
- Zoological Research is recognized as a core journal by the Research Center of Chinese Science Evaluation (RCCSE)
- The internationalization voyage of Zoological Research

