Genome size of 14 species of fireflies (Insecta,Coleoptera, Lampyridae)
Gui-Chun Liu, Zhi-Wei Dong, Jin-Wu He,2, Ruo-Ping Zhao, Wen Wang, Xue-Yan Li,*, , , 650223,
2University of Chinese Academy of Sciences, Beijing 100049, China
3Center for Ecological and Environmental Sciences, Key Laboratory for Space Bioscience & Biotechnology, Northwestern Polytechnical University, Xi’an Shaanxi 710072, China
INTRODUCTION
Fireflies, in the family Lampyridae (Coleoptera), are well-known as luminescent insects and include more than 2 000 species in approximately 100 genera of seven subfamilies worldwide(Branham, 2010; Lawrence & Newton, 1995). Different firefly species and their developmental stages exhibit different signaling systems, which play important roles in sexual communication and defense. As such, fireflies are a good model for studying the evolution of luminous signaling systems (Stanger-Hall &Lloyd, 2015; Stanger-Hall et al., 2007), sexual selection, and speciation (Lewis & Cratsley, 2008; Lloyd, 1971,1973; Ohba,1983).
Eukaryotic genomes not only contain genetic information but also act as structural components that determine nuclear properties and influence various biological features such as cell size, developmental rate, and developmental complexity(Gregory & Hebert, 1999; Koshikawa et al., 2008). Genome size is described by either mass (pg) or number of base pairs(bp) (Gregory, 2005a). Eukaryotic genome size is important as the basis for comparative research into genome evolution and as an estimator of the cost and difficulty of genome sequencing programs for non-model organisms (Gregory, 2005b; Gregory et al., 2007).1
So far, the genome sizes of 5 635 animal species (3 793 vertebrates and 2 429 invertebrates) have been recorded in the Animal Genome Size Database (Accessed 27 March 2017)(Gregory, 2017). Compared to those of mammals (14.14%, 778 of 5 500 species) and birds (8.96%, 896 of 10 000 species), the genome sizes of invertebrates remain poorly studied regarding abundance and diversity. Of the nearly 1 000 000 described insect species, the genome sizes of only 930 (0.093%) have been estimated. Among them, more than two-thirds are from the Holometabolous orders Diptera (254 species), Coleoptera(181 species), Hymenoptera (153 species), and Lepidoptera(65 species) (Gregory, 2017). Coleoptera (beetles) (ca. 360 000 species) is the largest order in the animal kingdom(Bouchard et al., 2011, 2009), and its 181 species with reported genome size estimates are mainly distributed in nine families (Tenebrionidae: 69; Chrysomelidae: 65; Coccinellidae:39; Dermestidae: 6; Scarabeidae: 3; Dytiscidae: 2; Carabidae:1; Geotrupidae: 1; Silvanidae: 1). For the luminous beetle family (Lampyridae), the genome sizes of 23 species from North America have been described recently (Lower et al.,2017). Here, we report on genome size estimations of 14 firefly species from China.
To explore firefly genome size evolution and estimation of the cost and difficulty of future genome sequencing programs, we performedC-value measurements for 14 firefly species (two genera in Lampyrinae, three genera in Luciolinae, and one genera in subfamilyincertae sedis) using flow cytometry.Although many methods for the estimation of genome size have been described, most genome size estimates in both animal and plant species estimations have been conducted using flow cytometry (Galbraith et al., 1983; Gregory et al., 2013; Hare &Johnston, 2011). We also constructed a phylogenetic tree of the 14 species using a mitochondrial cytochrome oxidase subunit 1(COI) gene fragment and discussed firefly genome size evolution in the phylogenetic context. The relationships of genome size to morphological traits such as body length, body width, antennal length, and eye width were also described.
MATERIALS AND METHODS
Sampling and observation of morphological characteristics Specimens of 14 firefly species from Yunnan, Hainan, and Hubei provinces of China were used for genome size estimation and body size measurement (Table 1). Some live specimens were used for estimation of genome size, with the remaining samples kept in 75% alcohol for morphological observation and body size measurement. All morphological observations and measurements were carried out under a dissecting microscope(SMZ 800, Nikon, Japan) according to Jeng et al.(2007). All measurements were based on male adults as females were difficult to collect. The abbreviations BL, BW, EL, ELW, PL, AL,and EYW represent body length, body width, elytral length,elytral width, pronotal length, antennal length, and eye width,respectively. BL is the sum of PL and EL (BL=PL+EL), BW is the greatest distance across the elytra, and EYW denotes thesmallest interocular width (measured horizontally). Male genitalia were also dissected and examined under a dissecting microscope to help with specimen identification. According to previous morphological descriptions (Ballantyne et al., 2013; Jeng et al.,2000), all species were at least assigned to genus. For the four species with both male and female samples, live specimens collected at the same locality and time were observed to mate.Combined with their morphology, we confirmed they were of the same species.
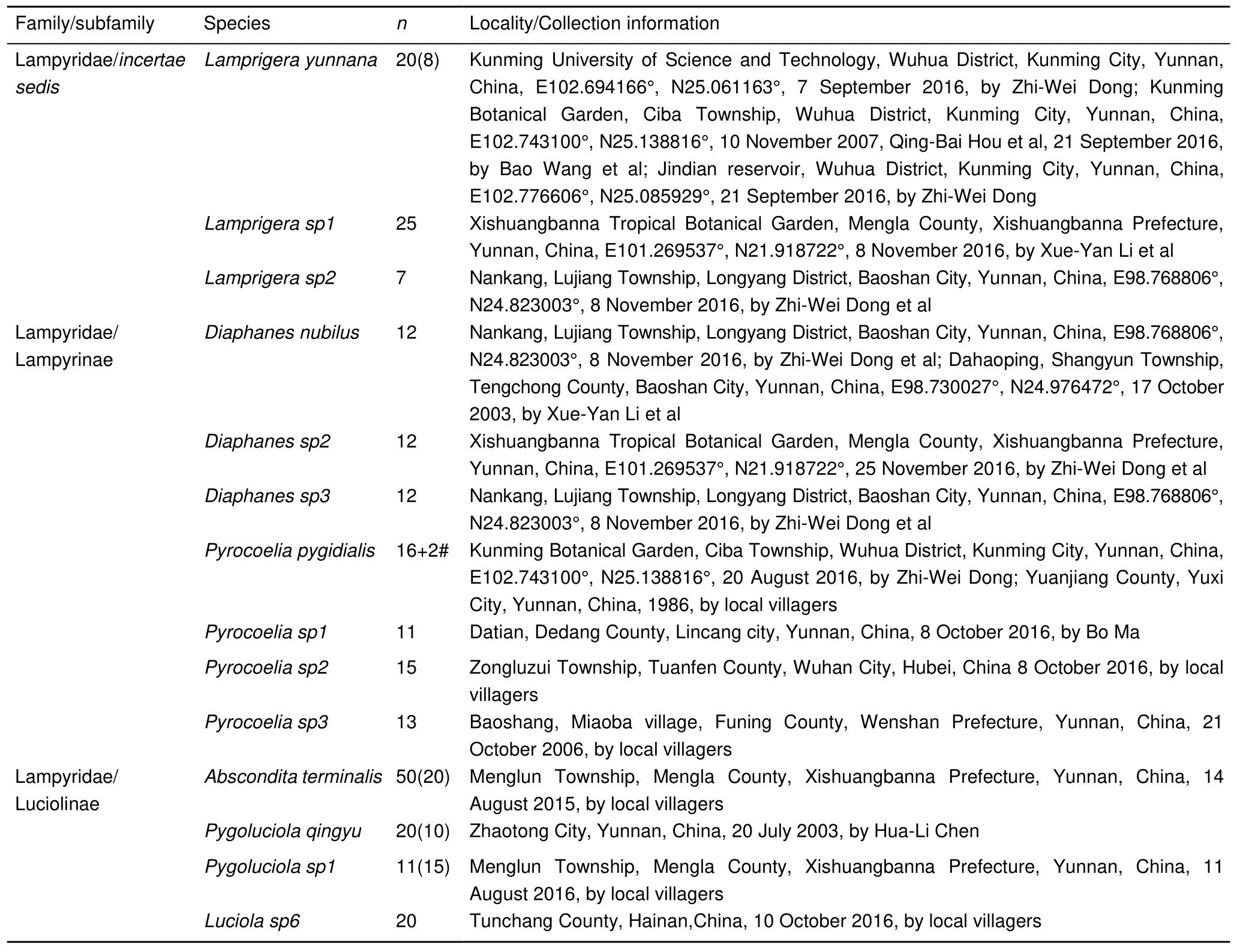
Table 1 Sample information in this study
For the males of each species, the brains of 3–6 live specimens were dissected for estimating genome size, with the thoraxes and abdomens were directly kept in –80 °C for genomic DNA extraction of single individuals when necessary. At least four males for each species were kept in 75% ethanol as voucher specimens. For females of the four species (Lamprigera yunnana,Abscondita terminalis,Pygoluciola qingyu, andPygoluciola sp1), brains of 4–6 live specimens were dissected to use for estimating genome size.
Flow cytometry
Genome size was estimated using flow cytometry (Bennett et al., 2003; Li et al., 2015). As with genome size estimation of other insects, such as the ladybird beetle (Gregory et al., 2003)and butterfly (Jiggins et al., 2005; Li et al., 2015), the model insectDrosophila melanogaster(genome size 176 Mb) (Bosco et al., 2007; Gregory & Johnston, 2008) was selected as the standard. Brain tissue from single firefly adults or larvae and the heads of 10Drosophila melanogaster(Dm) adults were dissected under a dissecting microscope (SMZ 800, Nikon,Japan) and added to 60 μL of cold Galbraith buffer (Galbraith et al., 1983) in 1.5 mL Eppendorf tubes in Pestles (Sigma, USA)issue grinder, stroked 40 times with a pestle, and then added to cold Galbraith buffer to get a final volume of 400 μL for Lampyridae and 1 000 μL forDm. Except forPyrocoelia pygidialis, we prepared cell suspensions from 3–6 males and 4–6 females of Lampyridae as biological replicates. ForP.pygidialis, only two larva individuals were used as biological replicates because no live adults were collected during the experimental period. Finally, theDmand firefly cell suspensions were filtered through a 20 μm nylon filter. After this, 50 μL of theDmcell suspension was added to 1.5 mL Eppendorf tubes containing 350 μL of the Lampyridae cell suspension. Propidium iodide was added to a final concentration of 50 parts per million,and the mixture was co-stained in the dark at 4 °C for 30–40 min. The fluorescence of co-stained nuclei for each sample was quantified using an LSR Fortessa (BD, USA) with the laser tuned at 561 nanometers. The DNA content (pg) was determined by comparing the ratio of the 2C mean of the tested samples with the 2C mean forDm(1C=0.18 pg) (Bennett et al.,2003; Galbraith et al., 1983). Genome size (bp) was calculated from DNA content (pg) following the formula (Dolezel et al.,2003): genome size (bp)=(0.978×109)×DNA content (pg).According to this formula, eachC-value was calculated based on the main peak of the 2C cells.
DNA extraction, PCR amplification, and sequencing
The genomic DNA of fireflies was obtained from the thorax and abdomen of a single male individual. DNA extractions were performed using a Gentra Puregene Blood Kit (Qiagen,Germany) following the manufacturer’s protocols. The primers C1-J-2183 (5'-CAACATTTATTTTGATTTTTTGG-3') and TL2-J-3014 (5'-TCCAATGCACTAATCTGCCATATTA-3') (Lower et al.,2017; Simon et al., 1994) were used for amplification of the a part (about 800 bp) of the mitochondrialCOIgene. The 20 μL reaction mixture consisted of 10 μL of 2×Trans Direct PCR SuperMix (Trans Direct Animal Tissue PCR Kit), 1 μL of forward primer (C1-J-2183) (10 μmol/L), 1 μL of reverse primer (TL2-J-3014) (10 μmol/L), 1 μL of DNA template, and 7 μL of ddH2O.The amplification protocol was as follows: initial denaturation and enzyme activation for 5 min at 94 °C, followed by 35 cycles for 30 s at 95 °C, 30 s at 55 °C, 60 s at 72 °C, with a final extension of 7 min at 72 °C, and 10 °C hold. The PCR products were electrophoresed using 1% agarose gel and sequenced by BioSune BiotechnologyCo., Ltd (ShangHai, China).. TheCOIsequences of seven species were from our firefly mitogenome project (MG200080–MG200086); and those of the other seven species were from the current study and were deposited in GenBank under accession numbers (MF375910–MF375916).
Phylogenetic analysis
All sequences were aligned using ClustalW and analyzed using MEGA 7.0 software (Kumar et al., 2016) and MrBayes version 3.1.2 (Huelsenbeck & Ronquist, 2001). Interspecific and intraspecific sequence divergences were calculated using the General Time Reversible (GTR+G+I) model with the pairwise deletion option in MEGA 7.0. Based on the GTR+G+I model,maximum likelihood (ML) tree was constructed using MEGA 7.0.Node supports for ML were inferred with bootstrap analysis(500 replicates). The Bayesian tree was established with MrBayes Version 3.1.2. The GTR+I+G model was selected via Modeltest version 3.7 and MCMC was run for 300 000 generations. The average standard deviation of split frequencies reached a value less than 0.01, with the Bayesian posterior probabilities calculated from the sample points after the MCMC algorithm started to converge (Zhan & Fu, 2011).Rhagophthalmus lufengensisandRhagophthalmus ohbai(GenBank accession No. DQ888607.1 and AB267275.1,respectively) were used as outgroups (Li et al., 2007). We used molecular phylogeny to correct for nonindependence of related species (Felsenstein, 1985; Lower et al., 2017).
Analysis of relationship between body size and genome size
Body size measurements, including BL, BW, AL, and EYW were determined based on 4–5 male individuals (Table 2). The relationships between genome size and body size were plotted using ggplot2 (Wickham, 2016). Phylogenetic generalized least squares (PGLS) in the R package nlme (Pinheiro et al., 2017)was used to analyze correlations between genome size and explanatory variables.
RESULTS
Firefly morphology
Considering that identification of fireflies at the species level is still unclear, especially for those species distributed in China,we assigned some specimens as speciesincertae sedis(sp) at a defined genus, and described their morphology (Figure 1,Table 2).Lamprigerawas placed in the subfamilyincertae sedis(Martin et al., 2017). Three species ofLamprigerahad similar outer shapes (Figure 1A–C), but could be separated by their genital morphology. Three species ofDiaphaneswere easily separated by their antennae (Figure 1D–F). Four species ofPyrocoeliawere separated by their wing and luminous organs(Figure 1G–J). Four species of Luciolinae were separated into three genera, includingAbscondita,Pygoluciola, andLuciolaby their wing, abdomen, luminous organs, and genitalia (Figure 1K–N).
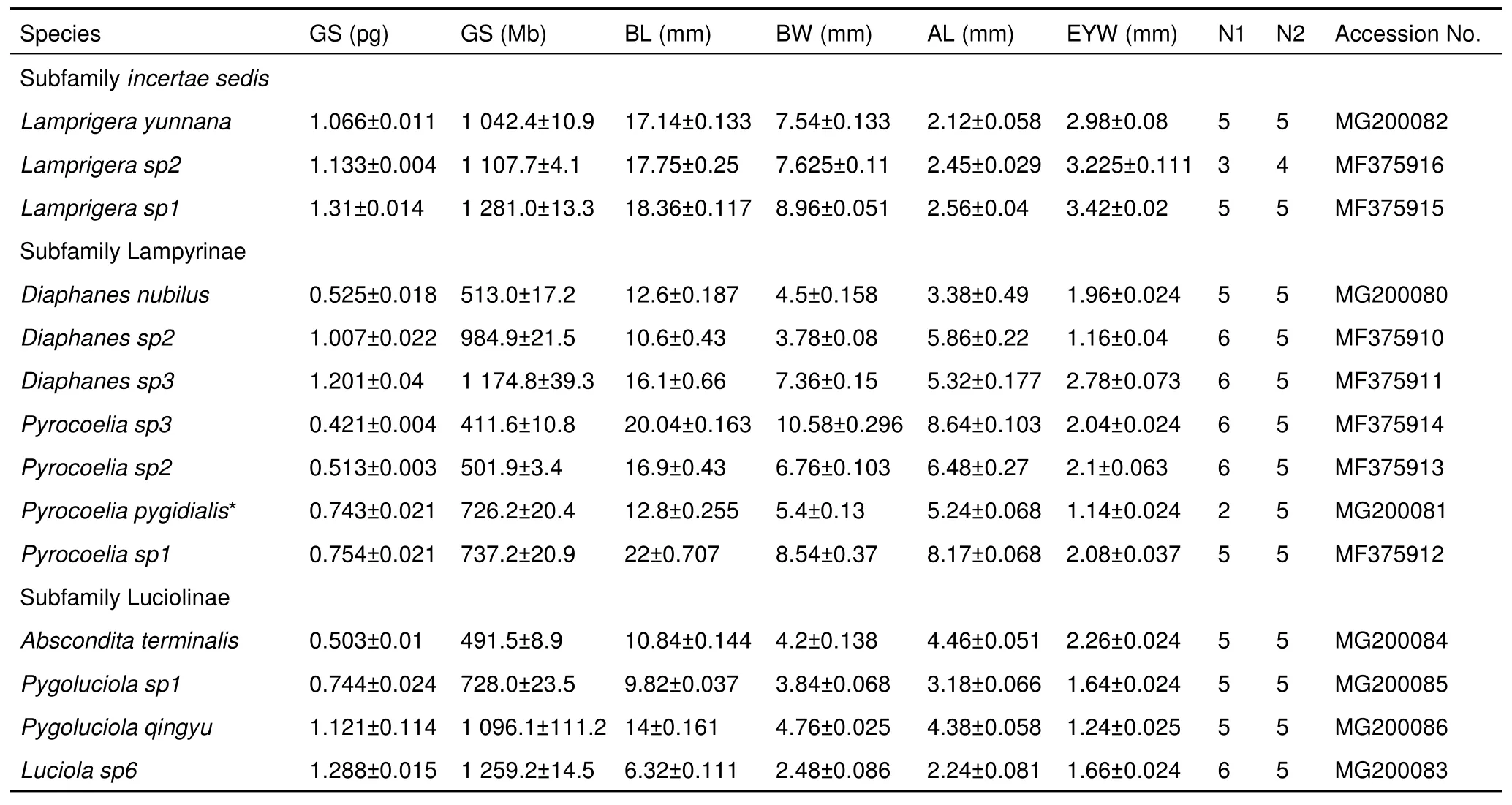
Table 2 Summary of the genome size (GS, in pg and Mb) of males of 14 firefly species and body size information, including body length(BL), body width (BW), antennal length (AL), and eye width (EYW)
Firefly genome size and evolution
Flow cytometry showed distinct peak(s) for the different species(Figure 2). Nuclei from the heads of the 10Dmspecimens and the brain of a singleLamprigera sp3male produced a single,broad 2C peak (Figure 2A–B), whereas mixtures of the heads ofD. melanogasterand brain of theLamprigerasp1male produced two broad 2C peaks (Figure 2C).
The haploid genome sizes of Lampyridae males ranged from 0.42 (Pyrocoelia sp3) to 1.31 pg (Lamprigera sp1) (411 Mb to 1 281 Mb) (Table 2), demonstrating 3.1-fold variation (Table 3).For four species:Lamprigera yunnana, Abscondita terminalis,Pygoluciola qingyu, Pygoluciola sp1, we also estimated the genome sizes of female individuals, which were found to be similar to those of the males (Table 4).
To explore the evolution of genome size within Lampyridae,we constructed a molecular phylogenetic tree for the tested species using the mitochondrialCOIsequences, which supported morphological taxonomy at the subfamily and genera levels (Table1, Figure 3).
Relationship between genome size and body size in fireflies
We explored the relationships between genome size and body size measurements, including BL, BW, AL, and EYW (Table 2).Our data showed no significant associations between firefly genome size and BL (r2=0.011,P=0.726,λ=1), BW (r2=0.016,P=0.669,λ=1), EYW (r2=0.11,P=0.241,λ=1), and AL (r2=0.045,P=0.469,λ=0.996) (Figure 4). We further performed PGLS analysis between BL, AL, EYW and phylogeny. The parameters of AL, EYW (λ=1), and BW (λ=0.996) indicated complete dependence on genome size between phylogeny and morphological traits. Pagel’s parameter estimates for genome size supported a Brownian motion model of evolution and complete phylogenetic dependence (λ=1.00, 95%) supported a neutral model (Lower et al., 2017).
DISCUSSION
Based on 39 species in 27 genera, the family Coccinellidae shows a large 26-fold genome variation (0.19–5.02 pg) (Table 3), with a considerable 21.7-fold variation also detected in

Figure 1 Habitus of 14 firefly species (All figures show dorsal view on the left and ventral on the right)

Figure 2 Number of nuclei measured by propidium iodide fl uorescence PI(PMT4)-stained flow cytometry

Table 3 Comparison of genome size for fireflies (Lampyridae) and other beetle families with described genome size
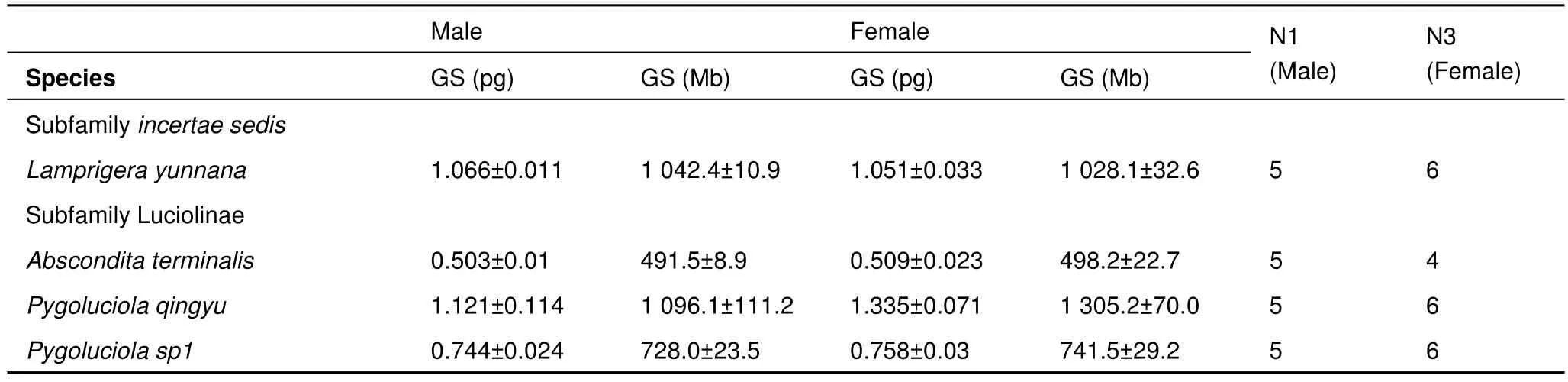
Table 4 Summary of genome sizes (GS, in pg and Mb) of males and females from four firefly species
Chrysomelidae (0.17–3.69 pg) according to 65 species in 27 genera (Gregory, 2017). A small 1.2-fold variation of genome size is reported in the family Dytiscidae (1.01–1.22 pg), though this is based on estimates of only two species. Our data from 14 species of six genera showed that the male haploid genome size in Lampyridae exhibited 3.1-fold variation (Table 3), which is relatively small compared to those of other currently estimated beetle families (Gregory, 2017) (Table 3).Nevertheless, compared to 2 000 species in more than 100 genera of seven subfamilies, the tested species in this study accounted for only a small proportion. Thus, more species,subfamilies, and genera, as well as different geographical distributions, are needed to better explore the evolution of firefly genomes. As Gregory (2002) states, theC-value enigma is a‘complex and multifaceted puzzle, immune to one dimensional explanations’.
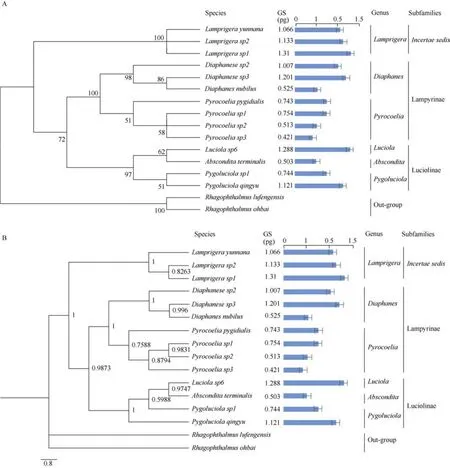
Figure 3 Phylogenetic trees of fireflies included in this study

Figure 4 Relationships between diploid genome size and body size (mm) in fireflies
Based on the phylogenetic relationship of the 14 species, our data suggest that genome sizes are very varied in Lampyridae.TheLamprigeraspecies in subfamilyincertae sedisexhibited a relatively large genome size of more than 1 pg (Table 2; Figure 3), which is less than 2-fold that of somePyrocoeliaspecies.The genome sizes of both Lampyrinae and Luciolinae ranged more than 2-fold. In Lampyrinae,Pyrocoeliaspecies had relatively small genomes, spanning 0.42–0.75 pg (411–737 Mb),including the smallest known genome (0.42 pg, 411 Mb) in Lampyridae (Table 2);Diaphanesspecies showed relatively large genome size variation, spanning from 0.53–1.2 pg (513–1 174 Mb), in whichDiaphanes sp2andDiaphanes nubilus,despite being closely related (Figure 3), showed 1.17-fold genome variation (Table 2). In Luciolinae, the genome sizes ofPygoluciola sp1 and Pygoluciola qingyuwere 0.74 pg (728 Mb)and 1.21 pg (1 096 Mb), respectively;Absconditaterminalishad a relatively small genome (0.5 pg, 491 Mb), but relatedLuciola(L. sp6) species had a large genome (1.29 pg, 1 259 Mb) (Table 2;Figure 3).
Except forLamprigera yunnana, three species in Luciolinae exhibited slightly larger genomes in females than in males.According to karyotypic analysis of species in the subfamilies Lampyrinae, Luciolinae, and Photurinae, Lampyridae frequently showed X0/XX karyotype sex determination, with males of X0 and females of XX (Dias et al., 2007), possibly explaining the slightly larger genome size in females than in males. Combined with the facts that the neoXY type was also reported from one species in Photurinae (Bicellonycha lividipennis) and the supernumerary chromosome found in some species of Lampyrinae (Dias et al., 2007) and thatLamprigerastill has a disputable position at the subfamily level (Jeng et al., 2000; Li et al., 2006), it is too early to explain the slight differences in genome size detected between males and females of this genera. Further karyotypic analyses of these genera should help to settle this question.
Our data showed no significant association between the firefly genome size and morphological traits such as BL, BW,and EYW (Figure 4). Previous data also support no correlation between genome size and body size in the beetle family Coccinellidae (Gregory et al., 2003) and in North American species (Lower et al., 2017). However, for thePimeliaandPhylangenera in the beetle family Tenebrionidae, negative correlations between genome size and body size have been reported (Palmer & Petitpierre, 1996; Palmer et al., 2003). For other insects such as aphids (Finston et al., 1995; Gokhman et al., 2017) and mosquitos (Ferrari & Rai, 1989) and other invertebrates such as turbellarian flatworms (Finston et al., 1995)and copepods (Gregory et al., 2000), a positive relationship between body size and genome size has been described.
Although the study of animal genome size has been ongoing for more than half a century, there is still a need to estimate the genome sizes of more animal groups by flow cytometry and further explore the evolution of genome size. Fast though costly next-generation sequencing technology will provide a complementary role for genome surveys, including genome size and complexity (Li et al., 2015). In summary, our study provides an estimation of the cost and difficulty of genome sequencing programs for non-model organisms, and will help promote studies on firefly genome evolution.
ACKNOWLEDGEMENTS
We would like to thank the anonymous colleagues and villagers for help in collecting the firefly specimens used in this study. We also thank Lei Chen and Wei Liu for their comments on this manuscript
Ballantyne L, Fu X, Lambkin C, Jeng ML, Faust L, Wijekoon WMCD, Li D,Zhu T. 2013. Studies on South-east Asian fireflies:Abscondita, a new genus with details of life history, flashing patterns and behaviour ofAbs.chinensis(L.) andAbs.terminalis(Olivier) (Coleoptera: Lampyridae:Luciolinae.Zootaxa,3721: 1–48.
Bennett MD, Leitch IJ, Price HJ, Johnston JS. 2003. Comparisons withCaenorhabditis(~100 Mb) andDrosophila(~175 Mb) using flow cytometry show genome size in Arabidopsis to be ~157 Mb and thus ~25% larger than the Arabidopsis genome initiative estimate of ~125 Mb.Annals of Botany,91(5): 547–557.
Bosco G, Campbell P, Leiva-Neto JT, Markow TA. 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species.Genetics,177(3): 1277–1290.
Bouchard P, Grebennikov VV, Smith ABT, Douglas H. 2009. Biodiversity of coleoptera.In: Foottit RG, Adler PH. Insect Biodiversity: Science and Society. Blackwell: Blackwell Publishing, 265–301.
Bouchard P, Bousquet Y, Davies AE, Alonso-Zarazaga MA, Lawrence JF,Lyal CH, Newton AF, Reid CA, Schmitt M, Slipiński SA, Smith AB. 2011.Family-group names in Coleoptera (Insecta).ZooKeys,(88): 1–972.
Branham MA. 2010. Lampyridae latreille, 1817.In: Leschen RAB, Beutel RG, Lawrence JF. Handbook of Zoology, vol IV, Arthropoda: Insecta,Teilband 39, Coleoptera, Beetles, vol 2, Morphology and Systematics.Berlin: Walter de Gruyter, 141–149.
Dias CM, Schneider MC, Rosa SP, Costa C, Cella DM. 2007. The first cytogenetic report of fireflies (Coleoptera, Lampyridae) from Brazilian fauna.Acta Zoologica,88(4): 309–316.
Dolezel J, Bartos J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human.Cytometry A,51(2): 127–128.
Felsenstein J. 1985. Phylogenies and the comparative method.American Naturalist,125(1): 1–15.
Ferrari JA, Rai KS. 1989. Phenotypic correlates of genome size variation inAedesalbopictus.Evolution: International Journal of Organic Evolution,43(4): 895–899.
Finston TL, Hebert PDN, Foottit RB. 1995. Genome size variation in aphids.Insect Biochemistry and Molecular Biology,25(2): 189–196.
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues.Science,220(4601): 1049–1051.
Gokhman VE, Kuhn KL, Woolley JB, Hopper KR. 2017. Variation in genome size and karyotype among closely related aphid parasitoids (Hymenoptera,Aphelinidae).Comparative Cytogenetics,11(1): 97–117.
Gregory TR, Hebert PDN. 1999. The modulation of DNA content: proximate causes and ultimate consequences.Genome Research,9(4): 317–324.
Gregory TR, Hebert PDN, Kolasa J. 2000. Evolutionary implications of the relationship between genome size and body size in flatworms and copepods.Heredity (Edinburgh),84(Pt 2): 201–208.
Gregory TR. 2002. Genome size and developmental complexity.Genetica,115(1): 131–146.
Gregory TR, Nedvěd O, Adamowicz SJ. 2003. C-value estimates for 31 species of ladybird beetles (Coleoptera: Coccinellidae).Hereditas,139(2):121–127.
Gregory TR. 2005a. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership.Annals of Botany,95(1): 133–146.Gregory TR. 2005b. The Evolution of the Genome. San Diego, CA: Elsevier.Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD. 2007. Eukaryotic genome size databases.Nucleic Acids Research,35(S): D332–D338.
Gregory TR, Johnston JS. 2008. Genome size diversity in the family Drosophilidae.Heredity (Edinb),101(3): 228–238.
Gregory TR, Nathwani P, Bonnett TR, Huber DPW. 2013. Sizing up arthropod genomes: an evaluation of the impact of environmental variation on genome size estimates by flow cytometry and the use of qPCR as a method of estimation.Genome,56(9): 505–510.
Gregory TR. 2017. Animal genome size database. http://www.genomesize.com. (Accessed March 27, 2017).
Hare EE, Johnston JS. 2011. Genome size determination using flow cytometry of propidium iodide-stained nuclei.Methods in Molecular Biology,772: 3–12.
Huelsenbeck JP, Ronquist F. 2001. MRBAYES: bayesian inference of phylogenetic trees.Bioinformatics,17(8): 754–755.
Jeng ML, Lai J, Yang PS, Sat? M. 2000. Notes on the taxonomy ofLamprigerayunnana(Fairmaire) and the genusLamprigeraMotschulsky(Coleoptera: Lampyridae).Japanese Journal of Systematic Entomology,6(2): 313–319.
Jeng ML, Yang PS, Engel MS. 2007. The firefly genusVestain Taiwan(Coleoptera: lampyridae).Journal of the Kansas Entomological Society,80(4): 265–280.
Jiggins CD, Mavarez J, Beltrán M, McMillan WO, Johnston JS, Bermingham E. 2005. A genetic linkage map of the mimetic butterflyHeliconiusmelpomene.Genetics,171(2): 557–570.
Koshikawa S, Miyazaki S, Cornette R, Matsumoto T, Miura T. 2008.Genome size of termites (Insecta, Dictyoptera, Isoptera) and wood roaches(Insecta, Dictyoptera, Cryptocercidae).Naturwissenschaften,95(9): 859–867.
Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets.Molecular Biology and Evolution,33(7): 1870–1874.
Lawrence JF, Newton AF. 1995. Families and subfamilies of Coleoptera(with selected genera, notes, references and data on family-group names).InPakaluk J, Slopinski SA. Biology, Phylogeny, and classification of Coleoptera: Papers Celebrating the 80th Birthday of Roy A. Crowson.Wilcza: Muzeum I Instytut zoologii Polska Akademia Nauk ul, 849–863.
Lewis SM, Cratsley CK. 2008. Flash signal evolution, mate choice, and predation in fireflies.Annual Review of Entomology,53: 293–321.
Li X, Yang S, Liang XC. 2006. Phylogeny of fireflies (Coleoptera: Lampyridae)inferred from mitochondrial 16S ribosomal DNA, with references to morphological and ethological traits. Progress in Natural sciences. 16 (8):817–826
Li XY, Ogoh K, Ohba N, Liang XC, Ohmiya Y. 2007. Mitochondrial genomes of two luminous beetles,RhagophthalmuslufengensisandR.ohbai(Arthropoda, Insecta, Coleoptera).Gene,392(1–2): 196–205.
Li XY, Fan DD, Zhang W, Liu GC, Zhang L, Zhao L, Fang XD, Chen L,Dong Y, Chen Y, Ding Y, Zhao RP, Feng MJ, Zhu YB, Feng Y, Jiang XT, Zhu DY, Xiang H, Feng XK, Li SC, Wang J, Zhang GJ, Kronforst MR, Wang W.2015. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies.Nature Communications,6: 8212.
Lloyd JE. 1971. Bioluminescent Communication in Insects.Annual Review of Entomology,16: 97–122.
Lloyd JE. 1973. Model for the mating protocol of synchronously flashing fireflies.Nature,245(5423): 268–270.
Lower SS, Johnston JS, Stanger-Hall KF, Hjelmen CE, Hanrahan SJ,Korunes K, Hall D. 2017. Genome size in North American fireflies:substantial variation likely driven by neutral processes.Genome Biology and Evolution,9(6): 1499–1512.
Martin GJ, Branham MA, Whiting MF, Bybee SM. 2017. Total evidence phylogeny and the evolution of adult bioluminescence in fireflies
(Coleoptera: Lampyridae).Molecular Phylogenetics and Evolution,107:564–575.
Ohba N. 1983. Studies on the communication system of Japanese fireflies.Science Report of Yokosuka City Museum,30: 1–62.
Palmer M, Petitpierre E. 1996. Relationship of genome size to body size inPhylan semicostatus(Coleoptera: Tenebrionidae).Annals of the Entomological Society of America,89(2): 221–225.
Palmer M, Petitpierre E, Pons J. 2003. Test of the correlation between body size and DNA content inPimelia(Coleoptera: Tenebrionidae) from the Canary Islands.European Journal of Entomology,100(1): 123–129.
Pinheiro J, Bates D, DebRoy S, Sarkar D, Eispack., Heisterkamp S, van Willigen B, R-core. 2017. nlme: linear and nonlinear mixed effects models.R package version: 3.1–131.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. 1994. Evolution,weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers.Annals of the Entomological Society of America,87(6): 651–701.
Stanger-Hall KF, Lloyd JE, Hillis DM. 2007. Phylogeny of North American fireflies (Coleoptera: Lampyridae): implications for the evolution of light signals.Molecular Phylogenetics and Evolution,45(1): 33–49.
Stanger-Hall KF, Lloyd JE. 2015. Flash signal evolution inPhotinusfireflies:character displacement and signal exploitation in a visual communication system.Evolution: International Journal of Organic Evolution,69(3): 666–682.
Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. 2nded.New York: Springer.
Zhan AB, Fu JZ. 2011. Past and present: phylogeography of theBufo gargarizansspecies complex inferred from multi-loci allele sequence and frequency data.Molecular Phylogenetics and Evolution,61(1): 136–148.
- Zoological Research的其它文章
- Obituary: Professor Colin Groves (1942–2017)
- Integrative taxonomy of Leptonetela spiders (Araneae,Leptonetidae), with descriptions of 46 new species
- Overview of the improvement of the ring-stage survival assay – a novel phenotypic assay for the detection of artemisinin-resistant Plasmodium falciparum

