Treatment repurposing for inflammatory bowel disease using literature-related discovery and innovation
Ronald Neil Kostoff, Michael Brandon Briggs, Darla Roye Shores
Abstract Inflammatory bowel disease (IBD) incidence has been increasing steadily, most dramatically in the Western developed countries. Treatment often includes lifelong immunosuppressive therapy and surgery. There is a critical need to reduce the burden of IBD and to discover medical therapies with better efficacy and fewer potential side-effects. Repurposing of treatments originally studied in other diseases with similar pathogenesis is less costly and time intensive than de novo drug discovery. This study used a treatment repurposing methodology, the literature-related discovery and innovation (LRDI) text mining system, to identify potential treatments (developed for non-IBD diseases) with sufficient promise for extrapolation to treatment of IBD. By searching for desirable patterns of twenty key biomarkers relevant to IBD (e.g., inflammation, reactive oxygen species, autophagy, barrier function), the LRDI-based query retrieved approximately 9500 records from Medline. The most recent 350 records were further analyzed for proof-of-concept. Approximately 18% (64/350) met the criteria for discovery (not previously studied in IBD human or animal models) and relevance for application to IBD treatment. Many of the treatments were compounds derived from herbal remedies, and the majority of treatments were being studied in cancer, diabetes, and central nervous system disease, such as depression and dementia. As further validation of the search strategy, the query identified ten treatments that have just recently begun testing in IBD models in the last three years. Literature-related discovery and innovation text mining contains a unique search strategy with tremendous potential to identify treatments for repurposing. A more comprehensive query with additional key biomarkers would have retrieved many thousands more records, further increasing the yield of IBD treatment repurposing discovery.
Key words: Treatment repurposing; Treatment repositioning; Inflammatory bowel disease; Literature-based discovery; Text mining; Crohn’s disease; Ulcerative colitis; Novel treatments
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal (GI) tract characterized by alternating phases of relapse and remission, clinically defined into two major subtypes: Crohn’s disease (CD) and ulcerative colitis (UC)[1]. Globally, the incidence of IBD is increasing, especially among newly industrialized countries where IBD was previously non-existent[2]. Childhood onset of IBD is also increasing in a similar pattern globally, suggesting evolving environmental risk factors[3].
In 2017, there were 6.8 million cases of IBD globally. The age-standardized prevalence rate increased from 79.5 per 100000 population in 1990 to 84.3 per 100000 population in 2017. At the regional level of global burden of disease (GBD), the highest age-standardized prevalence rate in 2017 occurred in North America (422.0 per 100000) and the lowest age-standardized prevalence rates were observed in the Caribbean (6.7 per 100000 population). High sociodemographic index (SDI) locations had the highest age-standardized prevalence rate, while low SDI regions had the lowest age-standardized prevalence rate[4].
The pathogenesis of IBD is multifactorial, which makes prevention and treatment of IBD challenging. Genetic predisposition contributes to dysregulation of both innate and adaptive immunity[5]. Environmental triggers (diet, infection, antibiotics, and toxin exposure) affect the intestinal microbiome and influence epigenetic changes that alter immune regulation[6]. Treatment, particularly in those with more aggressive disease, is generally lifelong, and often includes immunosuppressive therapy and surgery. There is a critical need to both (1) identify and eliminate contributing factors to disease to reduce the burden of IBD; and (2) discover novel medical therapies that provide better efficacy with fewer potential side-effects.
POTENTIAL IBD TREATMENT DISCOVERY
The standard approach to drug discovery is costly and time intensive. Repurposing treatments (1) already being developed; or (2) in clinical application for other conditions with overlapping pathogenesis is an attractive alternative tode novodrug discovery. This repurposing approach involves extrapolating a treatment developed for a non-IBD disease, such as rheumatoid arthritis, for use in IBD. Many of the upfront development costs can be avoided and some safety data already exists. However, given the vastness of the current published biomedical literature, quickly identifying potential novel treatments for IBD remains challenging.
A unique methodology to facilitate treatment repurposing has been developed by the first author using a literature-related discovery and innovation (LRDI) text-mining approach[7]. This approach involves matching patterns of biomarker changes (tests that measure a normal biologic, a pathologic process, a response to treatment, or predict a response[8]) in the disease-of-interest core literature with similar pattern changes in the remainder of the biomedical literature. The objective of this study is to use the LRDI approach to identify potential novel treatments for IBD that cannot be found in the IBD core literature.
However, the simultaneous removal of contributing factors to disease pathogenesis is equally important to treatment. A protocol (that includes identification and removal of contributing factors) to prevent and reverse chronic diseases is described in Appendix 1, along with examples of contributing factors associated with IBD. Contributing factors as used in the present study are, in theory, modifiable (reduced or eliminated through personal choice and/or government regulation), and can be broadly categorized as Lifestyle, Iatrogenic, Biotoxins, Occupational/Environmental, PsychoSocial/SocioEconomic. They include smoking, excessive alcohol, pesticides, wireless radiation, ionizing radiation, brominated flame retardants,etc[9].
METHODOLOGY
The specific details of LRDI-based treatment repurposing steps are contained in Appendix 2, as are the specific search terms used in the query. The query was executed April 19, 2020.
Figure 1 shows the flow diagram of the treatment discovery/repurposing process. Briefly, key biomarkers and their direction of change in the existing IBD literature were identified from examination of a large number of existing IBD treatments in all phases of development and clinical application (the reasons for identifying/using the large numbers of existing treatments are discussed in more detail in Appendix 3). These biomarkers and their desired treatment-derived directions of change are combined to form a query. The query was then used to search the non-IBD literature in Medline for potential treatments whose effects on biomarker changes were similar to those from existing IBD treatments in the core IBD literature (i.e., similar biomarker “signatures”). Those potential treatments from the non-IBD Medline literature that could not be found in the core IBD Medline literature were considered to be discovery.
Once the biomarkers associated with IBD were identified, they were ranked by (1) prevalence in the literature; and (2) clinical relevance. From hundreds of potential biomarkers, the top twenty (and variants) were prioritized and, along with their treatment-derived directions of change, were used in the treatment discovery query. Biomarkers from multiple pathways known to be associated with the inflammatory/immune response in IBD (i.e., cytokines, reactive oxygen species, autophagy and barrier function markers) were selected. All combinations of two biomarkers were then used to define the pattern for matching (e.g., decrease CRP AND increase IL-10). Categorical phrases, such as “anti-inflammatory” were also used as a biomarker term. The resultant query was entered into the Web of Science search engine for Medline.
In a proof-of-concept model, the 350 most recent records from 9500 records retrieved were analyzed further to (1) demonstrate the potential power of the technique; and (2) provide useful results to the IBD community as well. These potential treatments were then validated independently to assess applicability to IBD, in order to ensure the treatment was not already being tested in IBD models.
RESULTS
The treatments identified in this study include both (1) potential novel IBD treatments (Table 1)[10-73]; and (2) examples of recently studied IBD treatments that would have also been identified as potential novel treatments had this study been performed in 2016 (Table 2)[74-95]. The latter reflect the predictive nature of the LRDI treatment repurposing technique.
Table 1 contains the potential novel IBD treatments. The leftmost column contains the novel treatment and alternative name(s); the center column contains the biomarkers that were assessed in the reported study and that changed in the desired existing IBD treatment-derived directions; the rightmost column contains the reported study reference.

Table 1 Potential novel treatments

Biseokeaniamide A NO, iNOS, IL-1β, IkBα[48]Strigolactone GR24 NF-kB, Nrf2, PPARγ, occludin [49]Timosaponin BII MDA, GSH, PS, NLRP3, IL-1β, oxidative stress [50]Cycloastragenol (Y006)BAX, COX2, GSK3β, TNF-α, IFN-γ, IL-17, IL-10, IL-4[51]Ocellatin-K1(1-16); Ocellatin-K1(1-21)Nitrite, MDA, SOD, GSH, ROS, NF-kB, oxidative stress [52]Leocarpinolide B (LB)NO, PGE2, IL-6, TNF-α, MCP-1, COX-2, iNOS, NF-kB, ROS, HO-1, Nrf2[53]Enteromorpha powder GSH-Px, MDA, lipid peroxidation [54]Aminooxyacetic acid (AOAA)ATP, IL-6, TNF-α, IL-10, NLRP3, caspase-1, IL-1β[55]Gastrodin ROS, 8-OHDG, MDA, GSH-Px, SOD, Nrf2, HO-1, Bcl-2, Bax, caspase-3[56]Cinnamtannin D1 (CTD-1)IL-17, IL-6, IL-1β, TGF-β, IL-10, Th17, Treg, STAT5/Foxp3[57]ent-Kaur-15-en-17-al-18-oic acid ROS, MDA, GSH, SOD, NF-kB, bcl-2, p53, Bax, caspase-3[58]Hederacoside-C (HDC)IL-6, IL-1β, TNF-α, IL-10, TLR2, TLR4, MAPKs, NF-kB [59]PS-1145 dihydrochloride IL-6, TNF-α, IL-1β, NF-kB, COX-2[60]Cytokine-induced apoptosis inhibitor 1 (CIAPIN1)ROS, MAPKs, NF-kB, Bax, caspase-3, COX-2, iNOS, IL-6, TNF-α[61]Ruscogenin CRP, TNF-α, IL-6, IL-1β, ICAM-1, NF-kB, NOS-1[62]Phascolosoma esculenta IL-1β, TNF-α, IL-10, MDA, Nrf2, inflammation, oxidation [63]ALA/SFC Anti-oxidation, RANKL, IL-6, Nrf2[64]Xanthoplanine Inflammatory cytokines, ROS, STAT5[65]Lixisenatide ROS, NADPH, NOX-1, TNF-α, IL-6, IL-1β, MMP -2 -9, TLR4, NF-kB [66]Germanium MPO, TNF-α, IL-1β, IL-6, IL-10, NF-kB p65, p38, ERK, JNK [67]SDP Permeability, ZO-1, E-cadherin, NFkB, Il-6, hydrogen peroxide, IL-10[68]Amomum tsaoko IL-6, VEGF, Nh-kB, P-STAT3[69]Alkaline water ROS, SOD-1, GSH, telomerase activity, telomeres length [70]Dihydrotestosterone NO, PGE2, iNOS, COX-2, TNF-α, IL-1β, TLR4, NF-kB [71]Midazolam/Sufentanil TNF-α, IL-1β, HMGB1, NF-kB, ROS, SOD, inflammatory [72]JNJ16259685 Permeability, VASP, p-VASP, occludin, AQP [73]
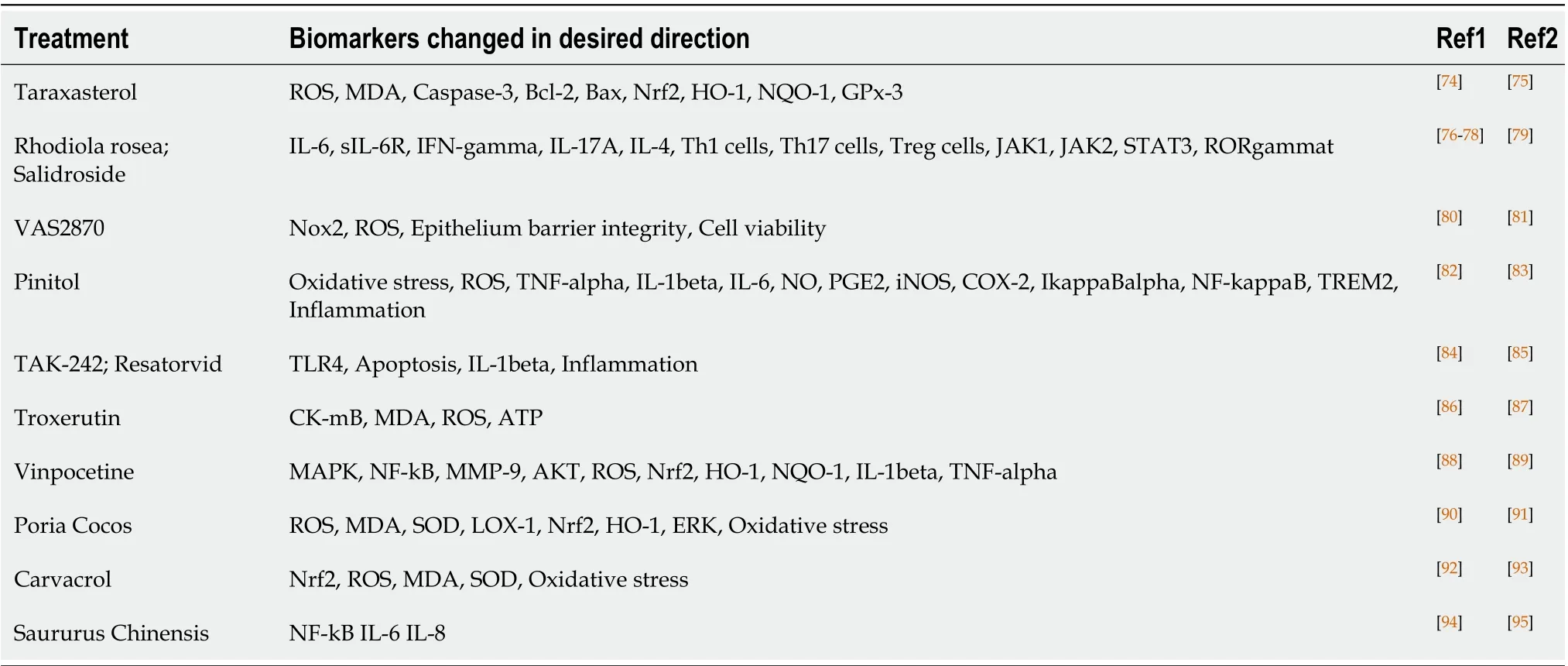
Table 2 Recently identified potential inflammatory bowel disease treatments.
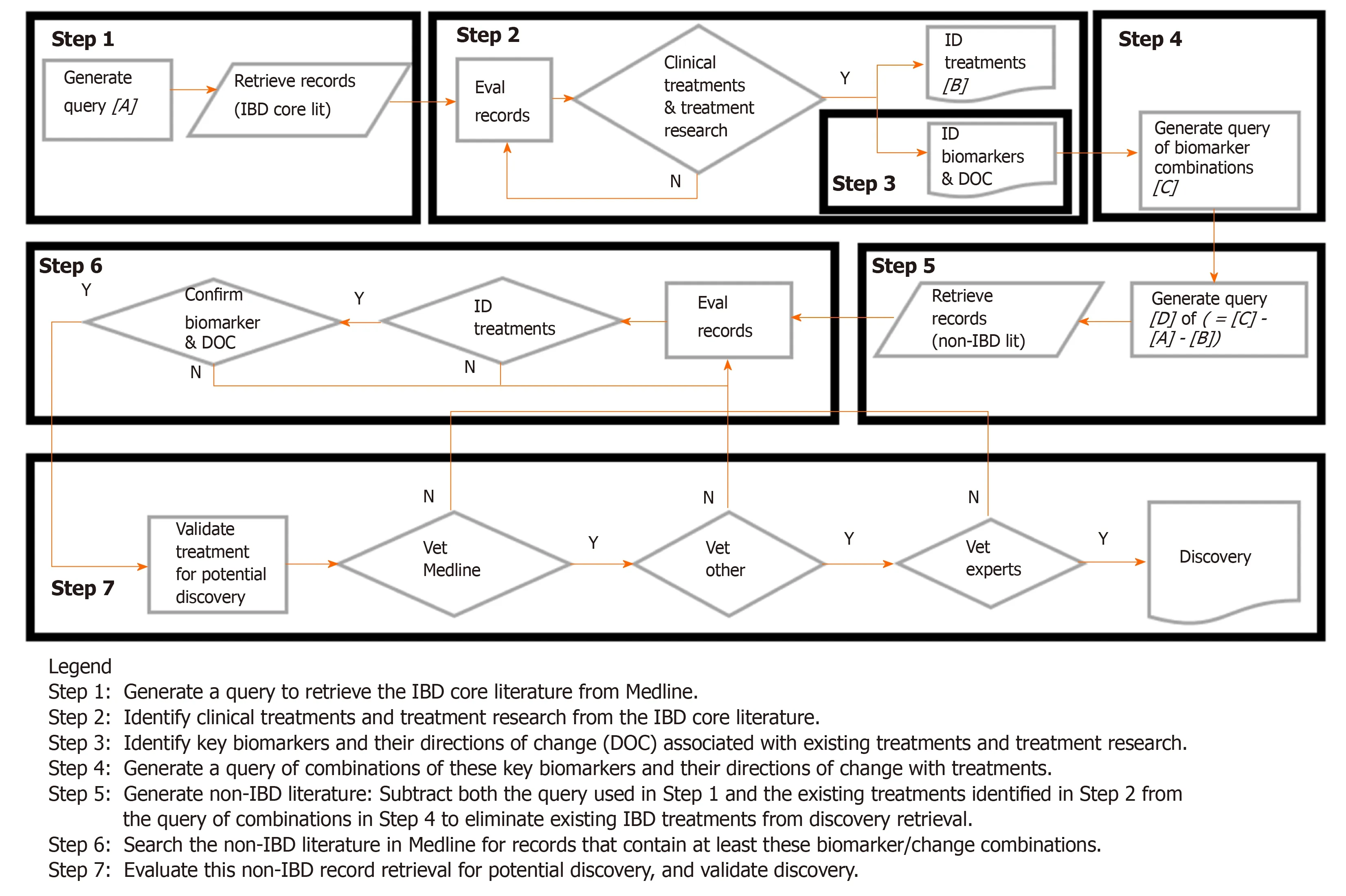
Figure 1 Literature-related discovery and innovation treatment repurposing flow diagram.
Table 2 shows examples of recent studies that would have been captured as novel treatments had the study/query been performed in 2016. These results reflect the predictive nature of the LRDI-based treatment repurposing methodology. The leftmost column contains the treatment and alternate name(s); the next column contains the biomarkers that were (1) assessed in the reported study; and that (2) changed in the desired treatment-derived direction; the final two columns reflect (1) the non-IBD application(s) of the treatment prior to 2016 (R1); and (2) the post-2016 evaluation of the potential treatment for IBD application (R2).
DISCUSSION OF RESULTS
The potential IBD treatments were derived from a wide swath of categories, including treatments for diabetes, cancer, and central nervous system disease, such as depression and dementia. Many of the compounds are derived from herbal remedies. Most are in the laboratory test phase, although many of these and their parent compounds have been in medical use for diseases other than the projected application represented by the paper cited. There is more emphasis on plant-based compounds and their derivatives than standard commercial synthetic drugs. This is to be expected, since the pharmaceutical companies who develop these drugs have substantial resources to devote towards searching for other applications for drugs under patent coverage with established manufacturing techniques.
Much of the research on plant-based treatments tends to be conducted by researchers indigenous to where these plants are most plentiful. They would not be expected to have the resources available to devote towards searching for the full spectrum of applications that the pharmaceutical companies have. Therefore, more targets of opportunity for treatment discovery exist in these non-mainline categories, as reflected in the results presented in this paper. It should be emphasized these drug/non-drug conclusions are based on results obtained using twenty selected biomarkers, out of a potential pool of hundreds of biomarkers. It is unknown whether the use of other available biomarkers for the discovery query would have generated novel treatments with a different drug/non-drug balance.
Approximately eighteen percent of the 350 records examined could be categorized as potential novel treatments for repurposing. Many of the records excluded from discovery consideration were done so on the basis of one or two prior recent records (published in 2017-2020) containing the same treatment applied to IBD (human or animal studies).
Thus, had this study been done four-five years ago, using the same query, those studies would have been identified as a potential novel treatment discovery. This is confirmation of the predictive power of the present technique for use as a repurposing approach.
CONCLUSION
Potential novel IBD treatments have been identified using a powerful text mining approach that identifies pattern changes of clinically relevant biomarkers being studied in non-IBD populations. Approximately 64 potential novel IBD treatments were identified in this proof-of-concept model, but many more were possible from an expanded study. Additionally, the predictive power of the approach for identifying future treatments that would be pursued through laboratory research was confirmed, showing the value of this approach for identifying and expanding relevant fields of research.
 World Journal of Gastroenterology2020年33期
World Journal of Gastroenterology2020年33期
- World Journal of Gastroenterology的其它文章
- Bowel function and quality of life after minimally invasive colectomy with D3 lymphadenectomy for rightsided colon adenocarcinoma
- Effects of denosumab treatment in chronic liver disease patients with osteoporosis
- Neutrophil to lymphocyte ratio and albumin bilirubin grade in hepatocellular carcinoma: A systematic review
- Radiomics of rectal cancer for predicting distant metastasis and overall survival
- Liver fat accumulation measured by high-speed T2-corrected multi-echo magnetic resonance spectroscopy can predict risk of cholelithiasis
- Exploring the food-gut axis in immunotherapy response of cancer patients
