Effects of purposeful soccer heading on circulating small extracellular vesicle concentration and cargo
Eri R.Muoz,Jlyn B.Cese,Brittny E.Wilson,Kyle T.Shuler,Fernndo V.Sntos,Crolin T.C'n,John J.Jek,Dinne Lngford,Mtthew B.Hudson,*
a Department of Kinesiology and Applied Physiology,University of Delaware,Newark,DE 19713,USA
b School of Health and Rehabilitation Sciences,The Ohio State University College of Medicine,Columbus,OH 43210,USA
c Department of Neuroscience,Lewis Katz School of Medicine,Temple University,Philadelphia,PA 19140,USA
Abstract
Keywords: Biomarker;Concussion;MicroRNA;Repetitive head impact exposure;Soccer heading
1. Introduction
Repetitive head impact exposure (RHIE) often does not result in acute clinical signs and symptoms of concussion but has the potential to cause significant detrimental neurological effects when experienced repeatedly.1The high prevalence of RHIE in sports (e.g., American football, ice hockey, and soccer) is concerning, given its possible association with acute and chronic neurological consequences, such as cognitive impairment,behavioral/mood dysfunction,and microstructural white matter brain changes.2-4Quantifying the manifestation of possible chronic effects is difficult because clinical measures lack the sensitivity to identify subtle changes in neurological functioning following RHIE. Furthermore, the possibility remains that null findings may, in fact, be attributed to the absence of neurological changes in the individual rather than the lack of sensitivity of the biomarkers. However, advanced neuroimaging (e.g., functional magnetic resonance imaging and diffusion tensor imaging) can identify functional impairments and changes in white matter microstructure but requires expensive equipment and advanced training.5,6In this context,other more sensitive and specific markers for acute and chronic changes are needed.
Small extracellular vesicles (sEVs) (<200 nm) are lipid vesicles released from most, if not all, cell types and contain a variety of molecular cargo including proteins, large RNAs,microRNAs (miRNAs), and DNA.7Recent evidence demonstrates that sEVs provide a stable and protective environment for molecular cargo to avoid degradation, making them attractive candidates for cell-type and biological response-specific biomarkers.8,9Several subpopulations of EVs have been identified,including microvesicles (100-1000 nm), and exosomes(30-120 nm).While each subpopulation differs in mechanism(s)of biogenesis and molecular contents,exosomes are the smallest and most widely studied.10Previous work suggests that the number of circulating sEVs increases following severe traumatic brain injury(TBI)in humans and a controlled cortical impact model in mice.11,12However, it is unknown if the concentration and/or size of sEVs in plasma changes with RHIE. In addition to concentration, altered expression of sEV cargo is implicated in a myriad of pathophysiologic processes,including neuroinflammation and neurodegenerative diseases.13-15
MiRNAs are small non-coding RNA molecules capable of targeting numerous mRNAs to inhibit translation or facilitate RNA degradation.16Altered concentrations of miRNAs are associated with numerous pathological conditions, including subconcussive trauma, concussion, and severe TBI.17-21We and others have shown that during injurious and pathological conditions, specific miRNAs are modified and selectively packaged into sEVs and released into the extracellular environment.22,23Specifically, we have demonstrated selective packaging of miR-182 and miR-23a into sEVs following muscle atrophy-inducing conditions. However, it is unknown if altered concentrations of miRNAs are associated with RHIE.Therefore, the purpose of this exploratory pilot study was to identify and investigate the miRNA profile in circulating sEVs derived from human plasma following RHIE. Since many sEVs released from cells enter systemic circulation, we hypothesized that even limited RHIE results in increased plasma sEV numbers and altered miRNA content. Furthermore, we hypothesized that alterations in miRNA content would be associated with neurodegenerative changes. Elucidating a circulating sEV miRNA profile could help to identify novel biomarkers for early detection and monitoring of neurotrauma.
2. Methods
2.1. Participants
The University of Delaware Institutional Review Board approved the study. Participants provided written informed consent.Samples obtained from 21 participants were analyzed in this study (Tables 1 and 2). All participants were currently active soccer players on a recreational, intramural, or club team, were between 18 and 35 years old, and had 5 or more years of soccer heading experience. Exclusionary criteria included any head,neck,face,or lower extremity injury in the past 6 months;a history of balance problems or vestibular dysfunction; current use of any medications affecting balance;any neurological disorder;and any unstable cardiac or pulmonary disease. Soccer goalkeepers were excluded because they do not participate in routine soccer heading drills.
2.2. Study design
This study used a repeated measures design across 2 time points, baseline (Pre), and 24-h post-intervention (Post-24h).While much is still unknown about the timeline of sEV biogenesis in response to head impact (HI), it is well-established the inflammatory response to head injury peaks over the course of hours to days.24-26Previous research indicates that it takes numerous hours to days from the time initial“damage”to cells or tissues occurs until “packaged sEVs” are present in the blood. In some pathologies and conditions, sEVs containing the packaged cargo of interest are present for several days after initial cell or tissue damage. Based on these studies and other published and preliminary evidence from our lab,we estimated that the 24-h post-head-impact would be the most likely time point to collect blood containing sEVs that were “selectively packaged” because of the impact. Participants were randomly assigned to one of 3 intervention groups: control (CON), HI,or leg impact (LI). The interventions (described in detail below) occurred immediately following the baseline blood draw.At both time points(Pre and Post-24h),16 mL of whole blood was drawn through venipuncture by a certified phlebotomist. Participants were scheduled at their convenience. Each participant was tested at approximately the same time each day(i.e.,morning,afternoon,evening),but this was not consistent across participants.Participants were not fasting prior to either blood draw. Participants were asked to refrain from alcohol and exercise prior to testing.
2.3. Interventions
The HI group performed a soccer heading paradigm,which was developed to serve as an in vivo model of functional HI testing in non-helmeted athletes27and has been used extensively in the past to investigate the biomechanics and effects of RHIE in sport.28-31Briefly, standard soccer balls (Size 5,450 g, inflated to 8 pounds per square inch (psi)) were projected from a JUGS soccer machine(JUGS Sports Inc.,Tualatin, OR, USA) traveling at 11.2 m/s and covering 12 m. The HI group performed 10 standing headers in 10 min;HI participants were instructed to head the ball back in the direction from where it was projected. The LI group performed 10 LIs in 10 min; LI participants were instructed to “trap” the ball with their gastrocnemius.The CON group did not perform any headers or LIs. All HI and LI participants were able to head the balls and trap the balls as instructed while being monitored for compliance by a member of the research team.

Table 1 Demographic information for each group(mean±SD or n).

Table 2 Demographic information for next-generation sequencing.
2.4. sEV isolation
Whole blood samples in buffered-sodium-citrated tubes were gently inverted several times and centrifuged, with the brake fully disengaged, at 2500 g for 15 min at 21?C. After centrifugation,plasma was collected,placed into a 15-mL falcon tube,and centrifuged at 2500 g for an additional 15 min at 21?C to remove platelets. The platelet-depleted supernatant was aliquoted into internally threaded cryovials,snap frozen in liquid nitrogen,and stored at-80?C until use.Following thawing, the plasma was centrifuged at 12,000 g for 45 min at 4?C to remove any remaining platelets and cellular debris. The supernatant was aliquoted and centrifuged at 20,000 g for 60 min at 4?C to pellet larger EVs. The supernatant was then aliquoted and centrifuged at 103,860 g for 3 h at 4?C to pellet sEVs. The sEV pellet was resuspended in sterile phosphatebuffered saline (PBS) (50 μL 1× PBS/1 mL plasma) and stored at-80?C until use.
2.5. Nanoparticle tracking analysis(NTA)
NTA was performed on the CON,HI,and LI groups at Pre and Post-24h (n=18, 6 CON, 6 HI, 6 LI) to confirm plasma sEV concentration and size following RHIE. A NanoSight NS300 (Malvern Panalytical, Malvern, UK) equipped with a 532-nm green laser and NS300 FCTP Gasket (Cat no.NTA4137; Malvern Panalytical) was utilized for characterizing the size and number of sEVs.Video capture settings(camera Level 12) were set to record in triplicate for 45 s each.Samples were diluted (1:20) in a total volume of 400 μL of sterile,filtered PBS and injected using a 1 mL sterile BD Plastipak syringe (Becton Dickinson S.A., Madrid, Spain). The size and number of plasma sEVs were compared across groups and time points. Data were analyzed using NTA 3.2.16 software(Malvern Panalytical)with a detection threshold of 4.
2.6. Next-generation sequencing(NGS)and bioinformatics
The miRNAs were isolated and purified from plasma sEVs using the Total RNA Isolation Kit from Norgen Biotek (Cat no.37500; Thorold, ON, Canada), as we have previously described.22,23NGS was performed on miRNA samples isolated from plasma sEVs of the HI group Pre and Post-24h (n=3) and completed by Applied Biological Material Inc. (Richmond, BC,Canada).Briefly,quality control of samples was assessed with the Agilent 2100 Bioanalyzer(Agilent Technologies,Santa Clara,CA,USA)(data not shown).Libraries were constructed with the NEB Next Small RNA Library Prep Set and quantitated with the quantitative polymerase chain reaction (qPCR) Kit (KAPA SYBR FAST,Kapa Biosystems,Wilmington,MA,USA)and sequenced on a NextSeq 500(Illumina,San Diego,CA,USA)at 15 million reads (1×75 bp SE) per sample. Bioinformatic analysis of raw(FASTQ) data was performed by an independent bioinformatics expert at the Wistar Institute (Philadelphia, PA, USA). All reads from 16 bp to 28 bp were aligned using Bowtie32against human miRNA sequences from the Sanger miRNA database miRBase,33allowing for 1 mismatch.Candidate miRNAs were determined by expression level changes with the EdgeR algorithm34using paired t tests,passing a false detection rate of less than 5%as a threshold.
2.7. qPCR
The miRNAs from the NGS panel were measured in the LI group Pre and Post-24h (n=6) via qPCR to verify specificity of HI.Following miRNA isolation and purification from sEVs,cDNA was synthesized from miRNA utilizing a qScript miRNA cDNA Synthesis Kit (Cat no. 89168-790; Quantabio,Beverly, MA, USA). Next, cDNA was further enriched using PerfeCTa PreAmp SuperMix (Cat no. 89409-170; Quantabio)to increase cDNA template volume for qPCR application.qPCR assays were then performed using primer sets designed to specifically amplify candidate miRNAs identified through NGS as the most changed in the HI group: miR-92b-5p,miR-423-5p, miR-24-3p, miR-7844-5p, miR-144-5p, miR-221-5p,and miR-22-3p.
2.8. TargetScan analysis and 3′-untranslated region luciferase reporter assay
To further explore the potential targets of miR-7844-5p, in silico target scan analyses were conducted. However, it is well-established that in silico miRNA target prediction is not always valid and accurate. Therefore, we utilized a luciferase reporter assay-based approach that we and others have previously described to validate specific miR-7844-5p mRNA targets from mRNAs of interest.22The cell-based screen was conducted using a custom panel of a total of 19 validated LightSwitch 3?untranslated region (UTR) GoClone reporter vectors(Active Motif,Carlsbad,CA,USA),including 6 previously established human endogenous 3?UTR GoClones,9 custom cloned 3?UTRs, a synthetic miR-7844-5p target as a positive control, and 3 controls including housekeeping controls and an empty vector. Human embryonic kidney (HEK)293T/17 cells (7×104) were co-transfected with a reporter vector(250 ng)and miR-7844-5p mimic(60 nM).After 24 h,the assay was developed with the LightSwitch Assay Reagent(SwitchGear Genomics, Menlo Park, CA, USA), and luminescence was analyzed using the average of 3 independent assays.
2.9. Statistical analysis
We compared demographic information between groups using one-way analysis of variance for continuous variables and Fisher’s exact test for count variables. Data are presented as mean±SE.A two-way repeated measures analysis of variance was utilized to compare the concentration and size of plasma sEVs from the NTA across groups (CON, HI, and LI)and time points (Pre and Post-24h). Paired t tests were used when comparing sEV miRNA concentrations from qPCR Pre vs.Post-24h in the LI group.Level of significance for all analyses was defined as a p value of less than 0.05.For age,height,weight,NTA,and qPCR comparisons,we report partial η2as a measure of effect size,whereby 0.01 is small,0.09 is medium,and 0.25 is large. For sex and race comparisons, we report Cramer’s V as a measure of effect size, whereby 0.1 is small,0.3 is medium, and more than 0.5 is large. Statistical analysis was conducted using GraphPad Prism 8.4.3 software(GraphPad,San Diego,CA,USA).
3. Results
3.1. RHIE does not affect plasma sEV concentration or size
To investigate the effect of RHIE on circulating sEV concentration and size we used NTA. Plasma sEV concentration(Fig. 1A) was not affected across time and group following RHIE(F=0.403,p=0.676,partial η2=0.051)(Table 3).Furthermore, plasma sEV size (Fig. 1B) was not affected across time and group (F=1.878, p=0.188, partial η2=0.200)(Table 3). Additionally, particle size distribution of plasma sEVs is presented in the CON (Fig. 1C), HI (Fig. 1D), and LI(Fig.1E)groups.Size distribution indicates the number of particles(concentration)within a given size(nm).
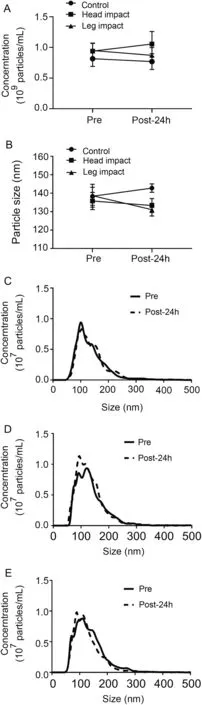
Fig.1. Repetitive head impact exposure does not affect plasma sEV isolation concentration and size. Plasma sEV concentration (A) and size (B) Pre and Post-24h.Plasma sEV size distribution for CON(C),HI(D),and LI(E)measured Pre and Post-24h.Data were analyzed by a two-way repeated measures analysis of variance and are represented by mean ± SE (A and B), n=18(6 CON,6 HI,6 LI).CON=control;HI=head impact;LI=leg impact;Pre=measured baseline;Post-24h=24-h post-intervention;sEV=small extracellular vesicle.
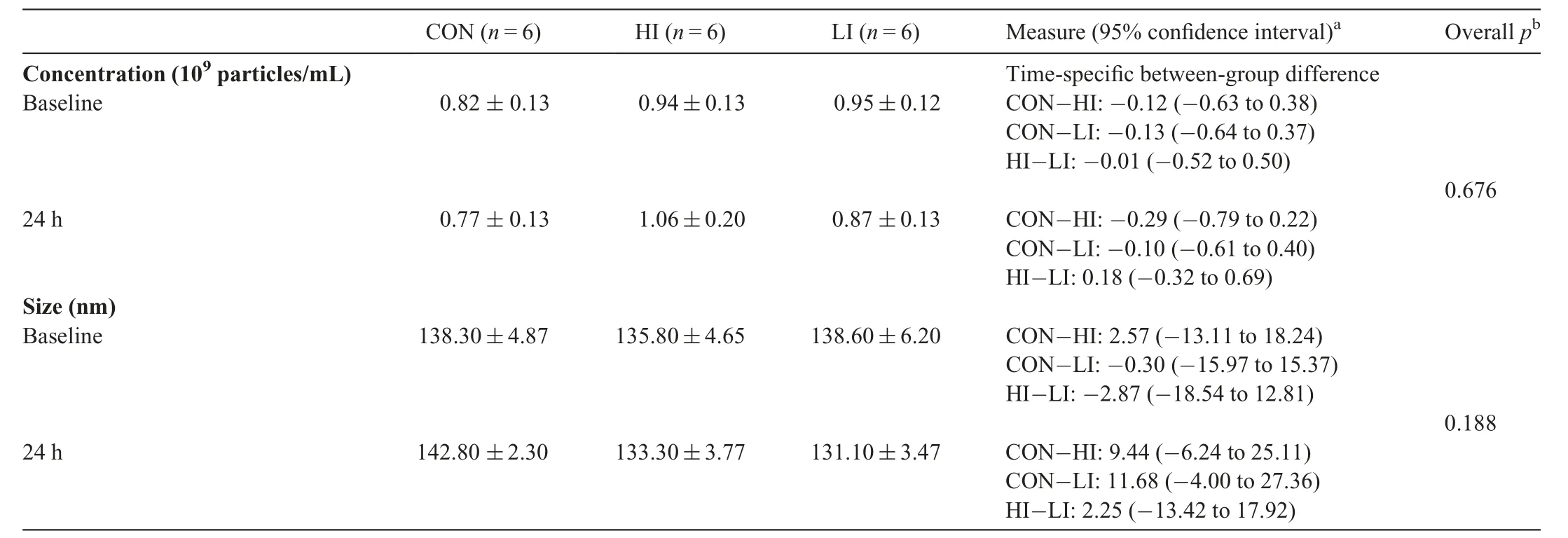
Table 3 Nanoparticle tracking analysis summary for each group(mean±SE).
3.2. RHIE results in miRNA signature in plasma sEVs
To determine the effect of RHIE on plasma sEV internal cargo, we utilized NGS to identify potential miRNA candidates. A miRNA signature in plasma sEVs was identified Post-24h, relative to Pre, in the HI group (Fig. 2). Based on abundance and fold change, a panel was identified including 7 miRNAs. NGS indicated 3 miRNAs increased more than 4-fold(miR-92b-5p,miR-423-5p,and miR-24-3p),and 4 miRNAs decreased more than 3-fold (miR-7844-5p, miR-144-5p,miR-221-5p, and miR-22-3p). The greatest change was observed in miR-7844-5p, which decreased by approximately 15-fold Post-24h(Fig.2).
3.3. LI does not affect plasma sEV miRNA signature concentrations
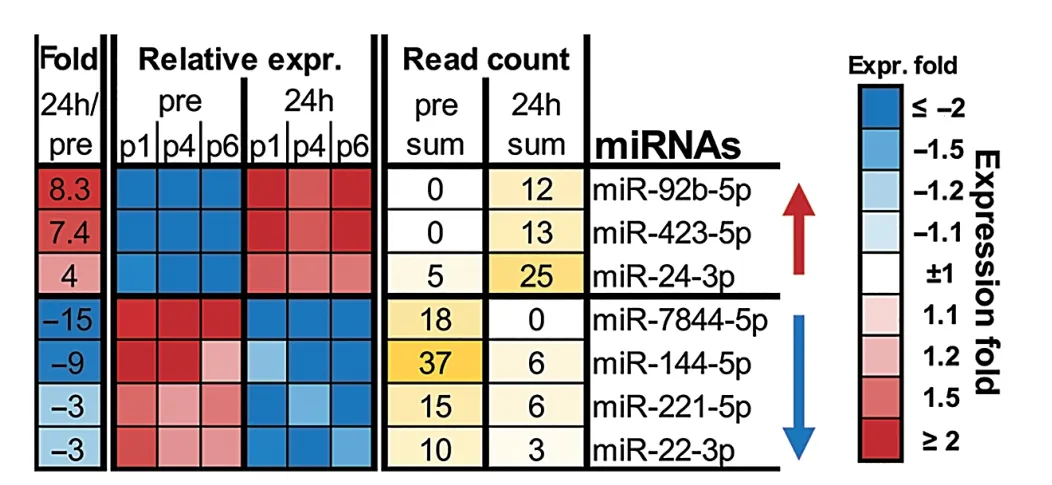
Fig. 2. Next-generation sequencing summary of the effect of repetitive head impact exposure on plasma sEV isolation miRNAs Pre and Post-24h. Seven candidate miRNAs were selected based on expression fold change(relative to Pre) using the EdgeR algorithm, n=3 (3 HI). expr.=expression; HI=head impact; Pre = measured baseline; Post-24h = 24-h post-intervention; sEV =small extracellular vesicle;miRNAs=microRNAs.
To determine if RHIE-associated miRNA changes were specific to HI,plasma sEV miRNA candidates were quantified via qPCR in the LI group (Fig. 3). No differences in the LI group were observed Pre vs.Post-24h(normalized to RNU48)for miR-92b-5p (p=0.622, partial η2=0.052), miR-423-5p(p=0.639, partial η2=0.047), miR-24-3p (p=0.650, partial η2=0.044), miR-7844-5p (p=0.288, partial η2=0.220),miR-144-5p (p=0.811, partial η2=0.013), miR-221-5p(p=0.761,partial η2=0.020),or miR-22-3p(p=0.242,partial η2=0.261)(Fig.3).
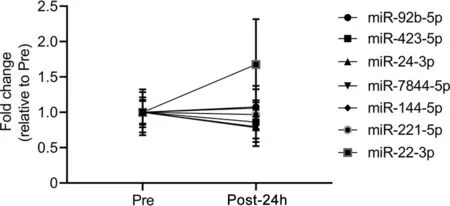
Fig.3. Leg impact (LI) does not affect plasma small extracellular vesicle isolation microRNA (miRNA) signature concentrations. The miRNA fold change relative to measured baseline (Pre) (normalized to RNU48) of LI is displayed for Pre and 24-h post-intervention (Post-24h) for miR-92b-5p, miR-423-5p,miR-24-3p, miR-7844-5p, miR-144-5p, miR-221-5p, and miR-22-3p. Data were analyzed by paired t tests and are represented by mean±SE,n=6(6 LI).
3.4. MiR-7844-5p targets mRNAs involved in mitochondrial apoptosis,autophagy regulation,mood disorders,and neurological dysfunction
To determine mRNA targets of miRNA-7844-5p, we used in silico target scan analyses and a luciferase reporter assay.The in silico target scan analyses indicated that miR-7844-5p was predicted to target mRNAs known to be involved in mitochondrial apoptosis, autophagy regulation, mood disorders,and neurological dysfunction (Fig. 4). Using a luciferase reporter assay-based approach, we validated 8 mRNA targets,including protein phosphatase 1 regulatory inhibitor subunit 1B(PPP1R1B);LIM and senescent cell antigen-like domains 1(LIMS1);autophagy-related 12(ATG12);microtubule-associated protein 1 light chain 3 beta (MAP1LC3B); integrin subunit alpha-1 (ITGA1); mitogen-activated protein kinase 1(MAPK1 V1, V2); glycogen synthase kinase 3β (GSK3β);and mitogen-activated protein kinase 8 (MAPK8) (Fig. 4).Conversely,analyses confirmed that the predicted mRNAs targets,including protein phosphatase 1 regulatory inhibitor subunit 14D(PPP1R14D),integrin subunit alpha-7(ITGA7),tight junction protein 1 (TJP1), stress-induced phosphoprotein 1(STIP1) homology and U-box containing protein 1 (STUB1),and occludin (OCLN), were not targets of miR-7844-5p(Fig.4).
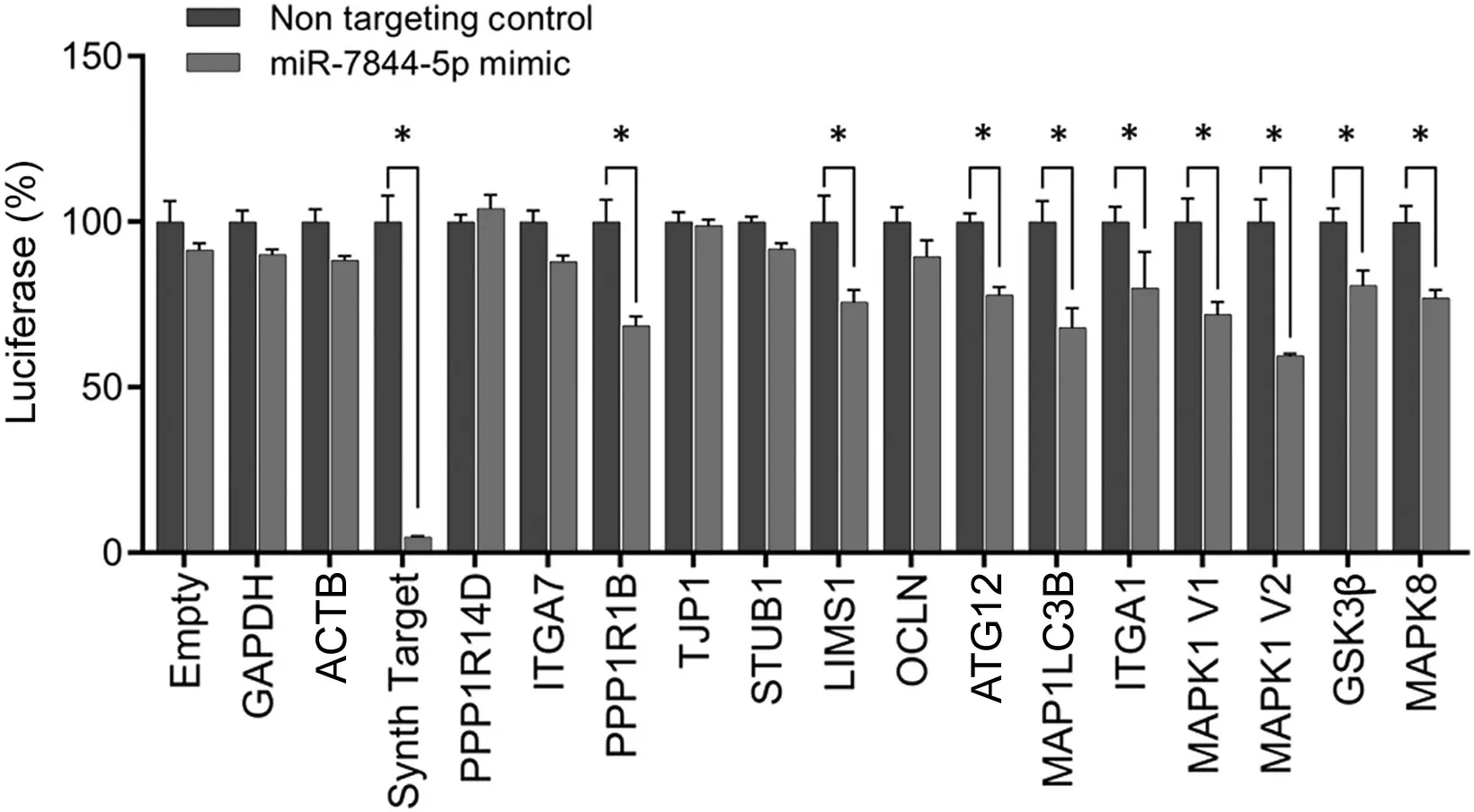
Fig. 4. LightSwitch luciferase reporter assay for miR-7844-5p. MiR-7844-5p was predicted to target 14 mRNAs known to be involved in mitochondrial apoptosis, autophagy regulation, mood disorders, and neurological disease. Eight mRNAs were validated as targets: protein phosphatase 1 regulatory inhibitor subunit 1B (PPP1R1B), LIM and senescent cell antigen-like domains 1(LIMS1), autophagy-related 12 (ATG12), microtubule-associated protein 1 light chain 3 beta (MAP1LC3B), integrin subunit alpha-1 (ITGA1), mitogenactivated protein kinase 1 (MAPK1) (V1 and V2), glycogen synthase kinase 3β (GSK3β), and mitogen-activated protein kinase 8 (MAPK8), n=3.*p <0.05. ACTB=beta-actin;GAPDH=glyceraldehyde 3-phosphate dehydrogenase;OCLN =occludin;PPP1R14D=protein phosphatase 1 regulatory inhibitor subunit 14D; STUB1 = stress-induced phosphoprotein 1 homology and U-box containing protein 1;TJP1=tight junction protein 1.
4. Discussion
The aim of this exploratory pilot study was to examine the feasibility of utilizing circulating sEV miRNAs as a biomarker of RHIE. Through routine contact sport participation, athletes experience RHIE, and some evidence indicates RHIE may be associated with long-term neurodegenerative disease (e.g.,chronic traumatic encephalopathy) development.35Clinical assessments may not be sensitive enough to identify the acute effects of RHIE, and more sensitive measures (e.g., neuroimaging) are too costly for common use. Although RHIE is not associated with acute physical, cognitive, or mental health impairments or with symptoms commonly associated with concussion,36RHIE may have accumulating effects over time.37,38A recent study showed that mortality rate from neurodegenerative diseases was significantly higher among Scottish former professional soccer players when compared to matched controls.39Former professional soccer players were also more likely to be prescribed dementia-related medication,while goalkeepers were among the least likely to receive a prescription for the treatment of dementia. These data highlight the importance of identifying a biomarker of RHIE to track the cumulative effects over time on players at risk of developing later-in-life brain health problems. Examining a biomarker capable of detecting RHIE may facilitate early detection and monitoring of future neurotrauma. Commercially available exosome isolation kits are becoming less expensive and easier to use, and thus sEV miRNAs are a potential feasible and affordable option for widespread use of biomarkers for disease manifestation and progression.
Previous studies have demonstrated that severe TBI and controlled cortical impact injury results in increased numbers of sEV in circulation;11,12however,in our study no changes in either concentration or size of sEVs were observed following RHIE.The exact mechanisms of TBI-induced increases in circulating sEV concentration are not completely understood.However,it is feasible that in our study the magnitude of HI in RHIE was not great enough to increase plasma sEV concentration. Next, we assessed cargo contained within sEVs, specifically miRNAs. NGS is a large-scale omics-based approach,and false-positive results can occur. However, as described in the Methods section, we limited the possibility of false-positive results by setting a false-positive detection rate of less than 5%. NGS revealed that, at Post-24h, fold changes for miR-92b-5p, miR-423-5p, and miR-24-3p significantly increased while fold changes for miR-7844-5p, miR-144-5p,miR-221-5p, and miR-22-3p decreased. Several studies have previously demonstrated that specific altered miRNA cargo following subconcussive and concussive trauma is associated with neurocognition, inflammation regulation, and neuronal repair.17-21In our study,the absence of any change in plasma sEV concentration combined with altered miRNAs following RHIE indicates that a selective miRNA packaging of sEVs took place.Furthermore,the absence of change in the level of these specific miRNAs in our LI group suggests that changes in the levels of these miRNAs did not occur due to general tissue damage or impact. Moreover, there are a number of impactful studies that have shown a close correlation between RNA sequencing data and qPCR results.40-42
Interestingly, several of these miRNAs have been previously been shown to be associated with neuropathology.For example,in some studies, miR-423-5p was associated with the progression of Alzheimer’s disease43-45and in our study, miR-423-5p was elevated in the HI group. MiR-22-3p (formally called miR-22),on the other hand,has been shown to protect against neurodegeneration,46and in our study its level decreased in our HI group. Lower levels of this miRNA have been associated with Alzheimer’s and Huntington’s disease.46Decreased levels of miR-144-5p have been associated with depressive symptoms,47and the level was lower in our HI group. Moreover, circulating miR-221 down-regulation, seen in our HI group, has been positively correlated with increased severity of Parkinson’s disease symptoms.48Last, miR-24-3p, which may promote glioma cell proliferation,49was elevated among those with acute concussion in one study,50and in our study was elevated in our HI group.Interestingly, several recent studies have postulated that neuroinflammation and brain trauma is a potential molecular mechanism in the development of glioblastoma.51,52Although these associations between miRNA and neurodegenerative diseases may be correlative, it is possible that sEV miRNA levels are indicative or predictive of future neuropathology.
The greatest miRNA change observed in our study was an approximately 15-fold decrease in miR-7844-5p Post-24h in the HI group.To our knowledge,this is a previously unidentified miRNA, and little to no current information exists about its role or function.In the present study we identified and validated numerous mRNA targets of miR-7844-5p (e.g.,PPP1R1B, LIMS1, AGT12), and these mRNA are known to be involved in mitochondrial apoptosis,53autophagy regulation,54mood disorders,55and neurodegenerative diseases.Mitochondrial dysfunction and impairments in autophagy have been linked as major causative factors in many neurodegenerative diseases, including Alzheimer’s disease,Parkinson’s disease, and amyotrophic lateral sclerosis.56,57Of special interest is LIMS1,which encodes for particularly interesting cysteine histidine-rich (PINCH) protein. Levels of PINCH protein increase during embryonic development, and the protein is involved in the regulation of cell migration and maintenance of neuronal polarity,58,59although it remains nearly undetectable in the CNS of healthy adults.60PINCH is ubiquitously expressed in numerous mammalian tissues,including the heart, kidneys, and liver. Dysregulation in PINCH expression has been reported in a range of diseases,including cardiomyopathy,61renal failure,62cancer,63and neurodegenerative disease.64PINCH is highly expressed in the brain tissue of patients with some neurodegenerative diseases,including Alzheimer’s disease, human immunodeficiency virus encephalitis, and frontotemporal dementia. Furthermore,PINCH has been identified as a potential contributing factor to the accumulation of hyperphosphorylated-tau, a hallmark of neurodegenerative tauopathies.Interestingly,silencing PINCH has significantly decreased hyperphosphorylated-tau in injured neurons.64In our study, we demonstrated that LIMS1 is negatively regulated, in part, at the translational level by miR-7844-5p.Therefore,we speculate that down-regulation of circulating miR-7844-5p following RHIE feasibly results in increased PINCH protein production and higher risk of accumulating hyperphosphorylated-tau during repeated neurotrauma. Conceivably, miR-7844-5p may have a neuroprotective role associated with PINCH and the accumulation of hyperphosphorylated-tau.
We acknowledge that there are several limitations to our study.In our controlled RHIE protocol,the HI group performed 10 soccer headers in 10 min. However, the number, frequency,and type of RHIE sustained during on-field participation may differ. Although our findings provide preliminary evidence of changes in sEV miRNAs following RHIE, sample sizes were low and NGS was limited to the HI group.In addition,although head acceleration,even during a controlled soccer heading paradigm, varies based on age, sex, neck strength, and other anthropometric measures, quantifying head kinematics was outside of the scope of this study. This soccer heading paradigm has been used in previous work to examine the biomechanics and effects of RHIE, but future work should examine the association between these sEV miRNAs and on-field RHIE in larger, more diverse cohorts and across more time points.
5. Conclusion
Altered sEV miRNAs may provide insight into the mechanisms involved in RHIE. Collectively, our data indicate that RHIE alters miRNA content of sEVs in circulation but does not alter the number or size of sEVs.
Acknowledgments
This study was supported by National Institutes of Health(NIH)-National Institute of Neurological Disorders and Stroke(NINDS) R01 (NS102157-01) and National Institutes of Health (NIH)-National Institute of General Medical Sciences(NIGMS)P20(GM113125-01).Graphical abstract and Video-Slides illustrations were created with BioRender.com.
Authors’contributions
ERM and JBC led the study design, data collection and interpretation, and drafting of the manuscript; BEW helped with data collection; KTS assisted with data collection and drafting of the manuscript; FVS participated in the study design and data collection; CTC aided in qPCR data collection; MBH, JJJ, and DL conceived the study and design,assisted with data interpretation, and provided a detailed review of the manuscript. All authors contributed to writing the manuscript and interpreting and analyzing the data. All authors have read and approved the final version of the manuscript,and agree with the order of presentation of the authors.
Competing interests
MBH is the founder and majority equity holder of the extracellular-vesicle-based company Extrave Bioscience, LLC.Extrave Bioscience, LLC., has licensed intellectual property from the University of Delaware related to some of the findings presented in the present study. MBH, JJJ, and TDL have separately filed for intellectual property licenses related to some of the findings presented in the present study through Temple University.All the support had no involvement in the study design and writing of the manuscript or the decision to submit it for publication.The authors declare that they have no other competing interests.
 Journal of Sport and Health Science2021年2期
Journal of Sport and Health Science2021年2期
- Journal of Sport and Health Science的其它文章
- Factors associated with concussion-symptom knowledge and attitudes toward concussion care seeking in a national survey of parents of middle-school children in the US
- The diagnostic and prognostic utility of the dual-task tandem gait test for pediatric concussion
- Impaired eye tracking is associated with symptom severity but not dynamic postural control in adolescents following concussion
- Slowed driving-reaction time following concussion-symptom resolution
- Impaired motor control after sport-related concussion could increase risk for musculoskeletal injury:Implications for clinical management and rehabilitation
- Detailed description of Division I ice hockey concussions:Findings from the NCAA and Department of Defense CARE Consortium
