Data Hiding in DNA for Authentication of Plant Variety Rights
Wei-Liang Tai,Charles C.N.Wang,Phillip C.Y.Sheu,and Jeffrey J.P.Tsai
1.Introduction
Living organisms have been excluded from patent laws for over two hundred years.Living organism forms were always considered as a product of nature and not a human invention in the past.Before 1930,plant breeders who created new types of plants had no claim to the marketing rights or sales of their plants,even though they have taken a lot of time and effort to breed a new plant.The non-patentable status of plants changed with the Plant Patent Act of 1930.According to the U.S.Department of Agriculture(USDA),the Plant Patent Act was enacted in 1930 and provided,for the first time,patent protection for new and distinct varieties of asexually reproduced plants[1].Now plant breeders have a financial incentive to perform plant breeding experiments and exercise control over their discoveries.
Although more than 68 countries protect new plant varieties by the plant breeder’s right,a few countries such as USA,European Union,Japan,and Australia do not exclude plant varieties from the patent right.The use of plant patents is becoming more and more common.In theory,molecular markers provide a method for helping to enforce plant patent rights.However,they are currently available for only a few breeders’ crop species since molecular marker libraries are expensive to develop.Furthermore,molecular markers represent a sample of the deoxyribose nucleic acid(DNA)from an organism rather than its entire genetic composition.Thus,molecular markers can easily demonstrate that two plants are different but hard to prove two plants are the same.For example,it may be difficult to find markers that differentiate a sport from the original cultivar,for the sport only differs from the original cultivar in a few genes.
Data hiding provide another efficient way for authentication and annotation of plant patents.Digital watermarking techniques have been proposed for extensive applications,such as ownership protection,copy control,annotation,and authentication.Most past work is designed for digital images where the pixels take on a wide range of colors or brightness levels.Nowadays,there are many plants are increasingly commonly seen in our life,such as rose,phalaenopsis,and vegetables.Having the capability of hiding data in DNA can facilitate the authentication,annotation,and tracking of these plants.However,hiding data in DNA is much more difficult than in digital images.For DNA sequences,the genetic codes take values from only four bases:adenine(A),guanine(G),cytosine(C),and thymine(T),and they are drastically different in functionalities.As a result,hiding data in DNA without changing the functionalities becomes more difficult.
Recently,hiding data in DNA becomes an interesting research topic because of biological property.In 2000,Leier et al.[2]proposed a special key sequence,called a primer,to decode an encrypted DNA sequence.They need to send a selected primer and an encrypted sequence to the receiver.Without the primers and the designated sequences,it is not possible to correctly decode the secret.Peterson[3]proposed a method to hide data in DNA sequences by substituting three consecutive bases as a character.However,there are at most 64 characters can be encoded in DNA.Further,high frequency of characters E and I can lead to the reveal of secret.
Chang et al.[4]proposed two schemes to hide data in DNA sequences with the reversibility.The first scheme compressed the decimal formatted DNA sequence using a lossless compression method and then appended the secret message to the end of the compression stream.Finally,the bit stream was converted back to nucleotides.The second scheme used the difference-expansion(DE)based reversible data hiding technique to conceal a secret bit in two neighboring words.In 2010,Shiu et al.[5]proposed three data hiding methods based upon properties of DNA sequences.These three methods are:the insertion method,the complementary pair method,and the substitution method.
Unfortunately,the robustness is low for some existing methods,which may result in inauthenticity and un-integrity of plant variety rights.Besides,these methods change not only the functionalities but also the original DNA sequence.Therefore,we propose a high capacity data hiding scheme for DNA without changing the functionalities of DNA sequences.This scheme adaptively varies the embedding process according to the amount of hidden data.
To make this paper self-contained,in Section 2,we describe the concepts from DNA that will be needed for the rest of the paper.Section 3 contains a detailed exposition of the proposed algorithm.In Section 4,we experimentally investigate the relationship between the capacity & hiding performance and the influence of variant DNA on the capacity.We also compare performance with existing schemes and analyze the robustness and security issues in this section.Finally,we conclude the paper in Section 5.
2.DNA
Recent advances and development in biological techniques research become more and more popular.In this section,we describe the DAN to functional protein called the central dogma as shown in Fig.1.The dogma is a framework for understanding the transfer of DNA sequence information.DNA is a molecule polymer of nucleotides that are collectively called the deoxyribose nucleic acid,which controls all biological life.The ribonucleic acid(RNA)is similar to DNA in structure,but in the fact that RNA contains the sugar ribose as opposed to “deoxyribose”.DNA has hydrogen at the second carbon atom on the ring,and RNA has hydrogen linked through an oxygen atom.
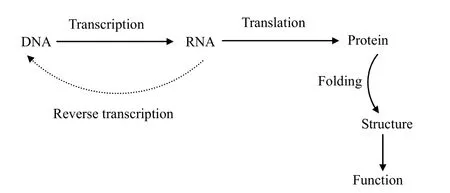
Fig.1.Central dogma of molecular biology.
In DNA and RNA,there are four nucleotide bases.Three of these bases are the same:A,G,and C.The fourth base for DNA is T,whereas in RNA,the fourth base lacks a methyl group and is called uracil(U).Each base has two points at which it can join covalently to two other bases on either end,forming a linear chain of monomers.
In general,RNA copies of DNA are made by a process known as “transcription”.Transcription is the conversion of information from DNA to RNA and is straightforward because of the direct correspondence between the four nucleotide bases of DNA and those of RNA.For most purposes,RNA can be regarded as the working copy of the DNA master template.There is usually one or a very small number of examples of DNA in the cell,whereas there are multiple copies of the transcribed RNA.The transcript RNA(tRNA)is referred to message RNA(mRNA)by discarding the intervening sequence of RNA.
In the protein-coding region of mRNA,three successive nucleotide bases,called triplets or codons,are used to code for each individual amino acid.Three bases are needed because there are 20 different types of amino acids shown in Table 1 but only four nucleotide bases:with one base there are four possible combinations; with two,16(42);with three,64(43),which is more than the number of amino acids.
The RNA transcription is used by a complex molecular machine called the ribosome to translate the order of successive codons into the corresponding order of amino acids.Special “stop” codons,such as UAA,UAG,and UGA,induce the ribosome to terminate the elongation of the polypeptide chain at a particular point.Similarly,the codon for the amino acid methionine is often used as the start signal for translation.
For plant variety rights,in the US the Plant Patent Act of 1930 initially allowed seed developers to take out patents on seeds they had developed by artificial selection.Now,the Act extends to the patenting of genetically modified(GM)crops by large multinational companies.It has succeeded in implementing similar patent laws in other countries.
Genetically modified crops have been around for over 20 years and have met with variable successes across the globe.Essentially,genetic modification involves inserting a section of DNA carrying a desirable gene of one species into the DNA of another.In crop plants,some of the beneficial features are achieved by using this method,including insect resistance,herbicide resistance(bar[6],aroA[7],bxn[8],sul,and tfdA[9]),nutritional value(OsNAS1,OsNAS2,and OsNAS3[10]),pest resistance,allergy resistance,higher yields,longer shelf life,and improved flavour.Therefore,in order to avoid altering DNA functionally,we propose a watermarking scheme based on mRNA sequences for copyright protection.
3.Proposed Scheme
An ideal watermarking system protects the data without altering it functionally.However,how can change DNA and still make the protein product come out the same? The only way is redundancy.Thus,we use the codon redundancy to hide data in mRNA sequence.A mRNA sequence is 3 bases long.There are 4 possible bases,U,C,A,and G.Clearly,64 unique codes can be generated.However,only 20 amino acids are encoded.There is redundancy in the mRNA to amino acid mapping.The redundancy can be used to hide data into the sequence without altering its function or length.
As shown in Table 1,there can be sets of 1,2,3,4 or 6 codons representing the same amino acid.Thus,we first take a binary secret sequence and convert it to a decimal number.Then we take the amino acid sequence that would be generated by the original mRNA,and generate a list of all possible codons that lead to the same sequence.The number of codon combinations for each amino acid is used to hide data.Having explained our background logic,we now outline the principle of the proposed data hiding algorithm.
3.1 Embedding Process
Input:a mRNA sequence with four possible bases,U,C,A,G,and a secret bit stream S={s1,s2,··,sn} where si∈{0,1} and 1≤i≤n.
Output:a watermarked mRNA sequence W={W'1,W'2,··,W'n} with the secret message S hidden.
Step 1.Translate the mRNA sequence into the amino acid sequence A={A1,A2,··,Am},which represents where Aiis a mRNA codon.
Step 2.Transform the secret bit stream to an unsigned integer B:
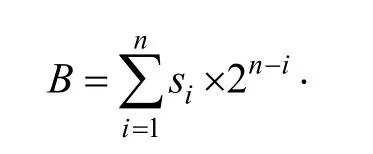
Step 3.Generate P={P1,P2,··,Pm} where Piis the number of codon combinations for each amino acid Aiin the sequence.
Step 4.Calculate the number T of all possible assignments for the input mRNA sequence based on the codon combinations until T>B:

where 1≤k≤m.
Step 5.According to the value of B,find the corresponding assignment of codons W'ifor all amino acids Ai,1≤i≤k,and output their codons:

Note that the number k should be used as a secret key for secure transmission.To show how this works,we take a sample message and embed it into a short sequence.Assume that the secret bit stream S={10011011} and the given mRNA sequence is “GCG CAG UUA GUA”.We first translate the mRNA sequence into the amino acid sequence A={Ala,Gln,Leu,Val}.The secret bit stream is transformed to an unsigned integer B=155.Then,we generate the number of codon combinations for each amino acid Aiin the sequence,P={4,2,6,4}.As shown in Step 4,we can obtain the number T=192 and k=4.Finally,as illustrated in Fig.2,we find the corresponding assignment of codons for all amino acids Ai,1≤i≤k,and output their codons.
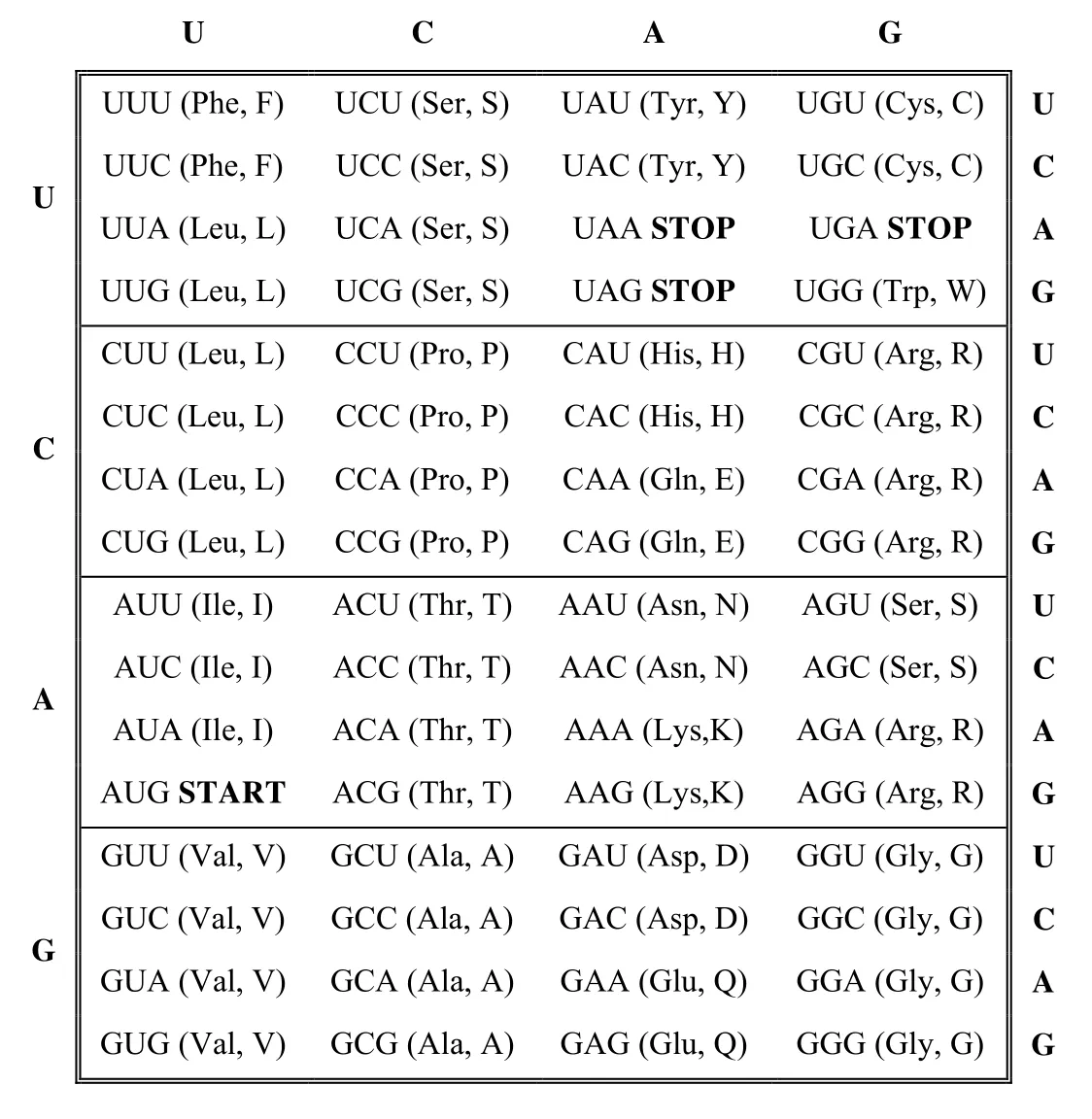
Table 1:Genetic code
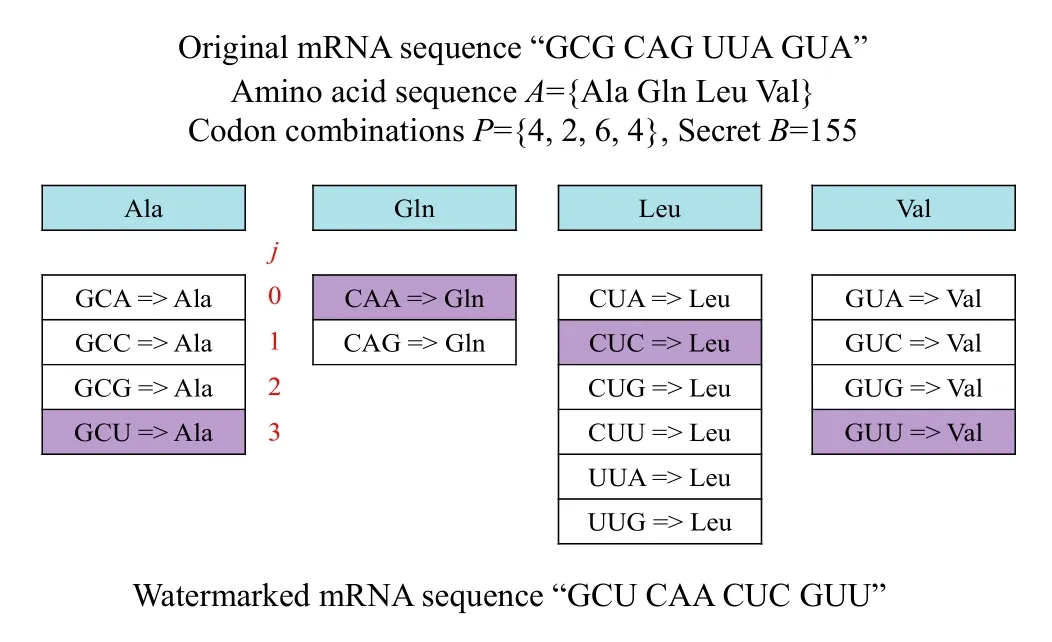
Fig.2.Embedding example.
This process has generated the new mRNA sequence“GCU CAA CUC GUU”.Therefore,following the highlighted path we get “GCU CAA CUC GUU”,which codes for the same amino acid sequence as the original mRNA,and also contains the hidden watermark.Note that there is adequate space to encrypt and embed a strong and repetitive watermark into living sequences since they consist of many thousands of base pairs.As a result,we use this technique in a retrovirus vector,that is,the message will transfer through the reverse transcription pathway and will be embedded in the DNA of the target cell in a complementary form.
3.2 Extraction Process
Input:a watermarked mRNA sequence and number k.
Output:secret message S.
Step 1.Calculate the series number B from the selected k codons.
Step 2.Transform the unsigned integer B to the secret bit stream si=(B/2n-i)mod 2,and output the bit stream.
The DNA sequence is deemed authentic only if the embedded authentication code matches the extracted information.
4.Experimental Results
To obtain a better understanding of how different host DNA sequences affect the performance of the proposed watermarking scheme,we present some results in a table form.All experiments were performed with eight significantly used genes:bar,aroA,BXN,sul,TfdA,OsNAS1,OsNAS2,and,OsNAS3,as shown in Table 2.These genes are commonly used for genetic modification in crop plants.Some of the beneficial features include insect resistance,herbicide resistance(bar[6],aroA[7],bxn[8],sul,and tfdA[9]),nutritional value(OsNAS1,OsNAS2,and OsNAS3[10]),pest resistance,allergy resistance,higher yields,longer shelf life,and improved flavour.The investment plants should be protected from any other acts that may be prescribed by the provisions of the Plant Varieties Act 1997.
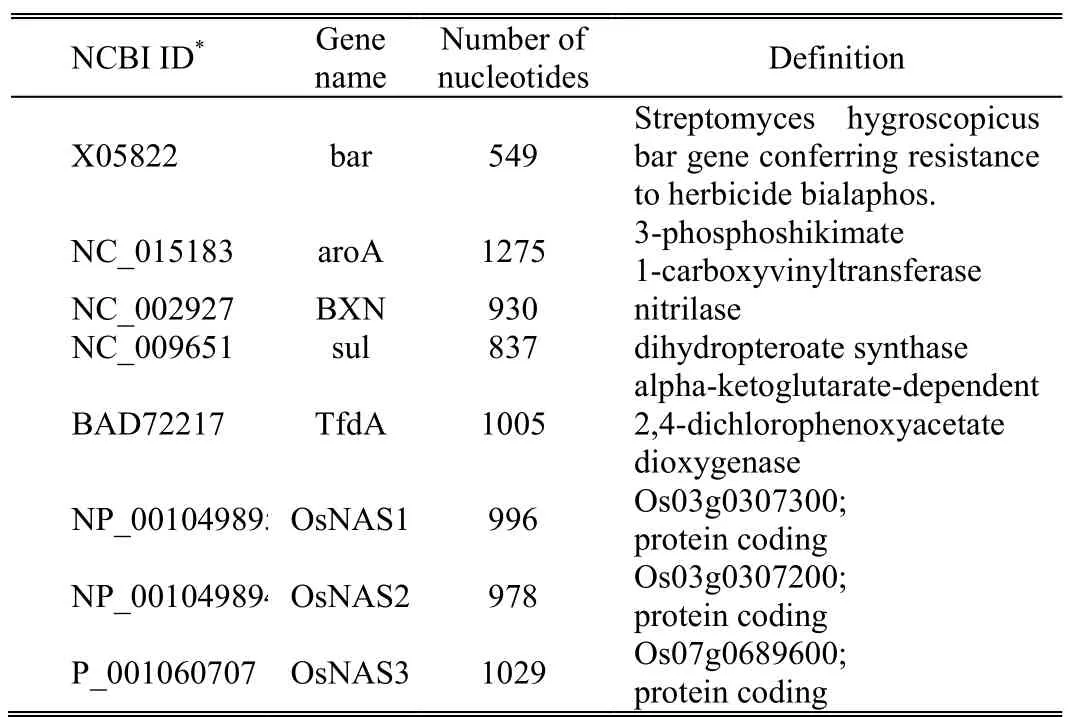
Table 2:Eight test DNA sequences
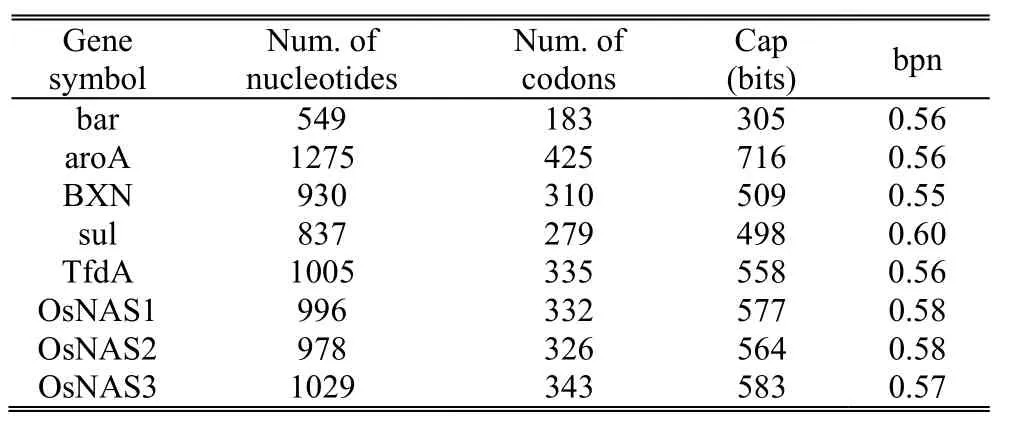
Table 3:Hiding performance of the proposed scheme
4.1 Capacity versus Distortion Performance
Table 3 gives an example of how different DNA sequences influence the payload Cap and the hiding performance bpn(bit per nucleotide),which is defined as

The real capacity that can be achieved depends on the nature of the DNA sequence itself.We notice that the average hiding performance is bpn=0.57.As a result,different DNA sequences do not influence the hiding performance.Our proposed scheme provides a stable hiding performance for authentication of plant variety rights.
4.2 Comparison with Other Schemes
Table 4 compares the hiding performance delivered by the proposed scheme and other existing schemes[4],[5].Note that the proposed scheme does not change the functionalities of DNA sequences,whereas schemes in [4]and [5]destroyed DNA sequences,which induces morphological changes in biological patterns.As Table 4 shows,the insertion method in [5]derived better performance.However,they did not preserve original characteristics of living organisms,which is unable to protect the copyright due to the functionalities change of genes.The evaluation results show that the proposed scheme achieves relatively high hiding performance than existing schemes.Further,our proposed scheme preserves the functionalities of genes,which ensure the copyright protection.

Table 4:Performance comparison with schemes[4],[5]
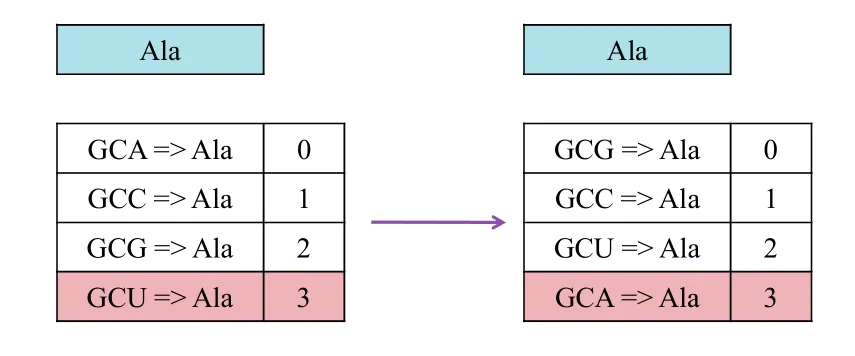
Fig.3.More robust variation in embedding process.
4.3 Robustness and Security
Security issues need to be handled by top layers in authentication applications.The major objective of an adversary is to forge authentication data so that an altered DNA can still pass authentication tests.Hence,it is important to study the following two problems for authentication applications:1)the probability of making DNA alterations while preserving the m-bit embedded authentication data,and 2)the possibility for an adversary to hide specific data in a DNA sequence.
In the mRNA sequence,there are 64 unique codes that can be generated in which there can be sets of 1,2,3,4 or 6 codons representing the same amino acid.In the proposed scheme,if a single codon is changed,the probability of getting a correctly decoded bit is
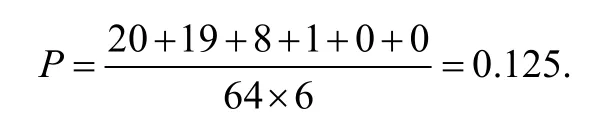
For the first issue,if the altered codons number is large,the probability of getting the decoded data to be exactly the same as the originally embedded one is approximately 2-3m,where m is the embedded payload size,which is very small as long as m is large.Therefore,the threat of making DNA alterations while preserving the m-bit embedded authentication data is very low.
The second issue depends on whether adversaries can derive information regarding the correspondence of codons and the carried bit by studying the difference between those copies,hence creating a new DNA sequence embedded with data when they will.Hence,assuming adversaries collect sufficient copies and know what data is embedded in each copy,they will be able to identify the correspondence of codons and the carried bit,and to hide their desired data by manipulating the corresponding codons.To prevent the above mentioned attack,we have to introduce more uncertainty.A good solution for dealing with this is to arrange the nucleotide randomly by using a secret key.As shown in Fig.3,instead of starting from the first codon chosen alphabetically as presented earlier,we can randomly number the codon before the watermark.This would result in a completely different sequence,but the information conveyed would be the same.This step keeps the process unambiguous,and at the same time ensures that it is difficult for an adversary to identify the congruent relationships of codons and the carried bits.
5.Conclusions
Hiding data in DNA can be applied to protecting the intellectual property in the gene therapy,transgenic crops,tissue cloning,and DNA computing.Data can also be hidden to verify the integrity of DNA sequences,authentication,and annotation for copyrights.Nowadays,molecular markers provide a method for helping to enforce copyrights.However,molecular markers can easily demonstrate that two plants are different but hard to prove two plants are the same.Therefore,we propose a watermarking scheme for authentication of DNA rights.To avoid changing the functionalities of genes,we embed data in the mRNA sequence by exploring the codon redundancy.We also address the robust and security issues and offer several additional steps to strengthen the watermark.As a result,the proposed scheme can actually be used to embed data directly into active genetic segments for copyright protection.In the other direction,one can generate an appropriate DNA sequence,when transcribed,which can create the mRNA strand with the embedded message.The practical usefulness of such a technique can be enormous.
[1]J.Chen,“The parable of the seeds:interpreting the plant variety protection act in furtherance of innovation policy,”Notre Dame Law Review,vol.81,no.4,pp.105-166,2006.
[2]A.Leier,C.Richter,W.Banzhaf,and H.Rauhe,“Cryptography with DNA binary strands,” BioSystems,vol.57,no.1,pp.13-22,2000.
[3]I.Peterson.Hiding in DNA.[Online].Available:http://www.maa.org/mathland/mathtrek_4_10_00.html
[4]C.-C.Chang,T.-C.Lu,Y.-F.Chang,and R.C.T.Lee,“Reversible data hiding schemes for deoxyribonucleic acid(DNA)medium,” Int.Journal of Innovative Computing,Information and Control,vol.3,no.5,pp.1-16,Oct.2007.
[5]H.-J.Shiu,K.-L.Ng,J.-F.Fang,R.C.T.Lee,and C.-H.Huang,“Data hiding methods based upon DNA sequences,”Information Sciences,vol.180,no.11,pp.2196-2208,Jun.2010.
[6]K.S.Rathore,V.K.Chowdhury,and T.K.Hodges,“Use of bar as a selectable marker gene and for the production of herbicide-resistant rice plants from protoplasts,” Plant Molecular Biology,vol.21,no.5,pp.871-884,Mar.1993.
[7]L.Comai,L.C.Sen,and D.M.Stalker,“An altered aroA gene product confers resistance to the herbicide glyphosate,”Science,vol.221,no.4608,pp.370-371,Jul.1983.
[8]D.M.Stalker,K.E.McBride,and L.D.Malyj,“Herbicide resistance in transgenic plants expressing a bacterial detoxification gene,” Science,vol.242,no.4877,pp.419-423,Oct.1988.
[9]J.F.Rice,F.M.Menn,A.G.Hay,J.Sanseverino,and G.S.Sayler,“Natural selection for 2,4,5-trichlorophenoxyacetic acid mineralizing bacteria in agent orange contaminated soil,” Biodegradation,vol.16,no.6,pp.501-512,Dec.2005.
[10]H.Inoue,K.Higuchi,M.Takahashi,H.Nakanishi,S.Mori,and H.K.Nishizawa,“Three rice nicotianamine synthase genes,OsNAS1,OsNAS2,and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron,” Plant Journal,vol.36,no.3,pp.366-381,Nov.2003.
 Journal of Electronic Science and Technology2013年1期
Journal of Electronic Science and Technology2013年1期
- Journal of Electronic Science and Technology的其它文章
- Design of MEMS C-Band Microstrip Antenna Array Based on High-Resistance Silicon for Intelligent Ammunition
- Low-Cost Embedded Controller for Complex Control Systems
- An Image Authentication Method by Grouping Most Significant Bits
- A Steganography-Based Optical Image Encryption System Using RGB Channel Integration
- Rejection of Direct Blast Interference Based on Signal Phase-Matching Array Processing
- Investigation on Active and Reactive Combined Spot Price Integrated with Wind Farm
