TaCML36,a wheat calmodulin-like protein,positively participates in an immune response to Rhizoctonia cerealis
Lin Lu,Wei Rong, Ronghua Zhou, Naxin Huo, Zengyan Zhang*
National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
Keywords:Calmodulin-like protein Defense-associated gene Ethylene signaling Sharp eyespot Triticum aestivum
ABSTRACT Sharp eyespot, mainly caused by the soil-borne fungus Rhizoctonia cerealis, affects wheat(Triticum aestivum L.) production worldwide. In this study, we isolated TaCML36 gene encoding a wheat calmodulin-like protein,and studied its defense role in protection against R. cerealis. Transcription of TaCML36 was significantly elevated by both R. cerealis infection and exogenous ethylene treatment.Transcription was higher in resistant wheat lines than in susceptible ones. There were copies of TaCML36 on chromosomes 5A, 5B, and 5D. The TaCML36 protein is composed of 183 amino acids and contains two calcium-binding EFhand domains.Subcellular localization assays in wheat indicated that TaCML36 localizes in both the cytoplasm and nucleus. Virus-induced gene silencing and disease assessment indicated that compared to the controls, TaCML36-silenced wheat plants displayed significantly reduced resistance to R. cerealis and had greater fungal biomass, suggesting that knockdown of TaCML36 impaired host resistance. Knockdown of TaCML36 also significantly repressed expression of pathogenesis-related genes such as Chitinase 1,PDF35, and PR17C, the ethylene response factor-encoding gene TaPIE1, and ethylene biosynthesis gene ACO2. Collectively, our results suggest that TaCML36 positively participates in the innate immune response to R. cerealis infection by modulating expression of defense-associated genes possibly in the ethylene signaling pathway.
1. Introduction
Common wheat (Triticum aestivum) is one of the most important staple food crops in the world. It provides 20% of the calorific intake of the human population. Thus, wheat production is essential for global food security [1]. Plant diseases not only inflict serious losses on wheat yields but also impact grain quality. Sharp eyespot, caused mainly by the necrotrophic fungus Rhizoctonia cerealis is a destructive disease of wheat worldwide [2-7]. R. cerealis infection can occur in the root and basal stem tissues at any time during the growing season. The disease can cause pre- and postemergence damping-off and shoot death of seedlings. Severe infection of mature shoots can cause small and shriveled grain, induce lodging at the second or third internodes and cause premature spike senescence or ripening (white heads)[7]. Since 2005, >6.67 million ha of wheat in China have been impacted annually by the disease [8,9]. In addition to sharp eyespot in wheat, R. cerealis also causes other diseases in important food and bioenergy crops,such as root rot in sugar beet, cotton, potato, and legumes [10,11]. Crop varieties with resistance are the most environmentally-friendly and effective method to control disease. Identification of major resistance-related genes and dissection of their functional roles are essential for the improvement of resistance to sharp eyespot.
Plants have sophisticated mechanisms to perceive and respond to environmental cues in order to survive and reproduce under adverse conditions [12]. Most external stimuli induce rapid changes in cytosolic calcium(Ca2+)levels that initiate signaling cascades to ultimately establish appropriate physiological responses[13].Ca2+signatures,as important secondary messengers, play key roles in developmental and growth processes, and in response to both biotic and abiotic stress [12,14-16]. To induce appropriate responses,these calcium messages are decoded, relayed, and amplified by Ca2+-binding proteins, termed Ca2+sensors. Calmodulin(CaM), which is conserved in all eukaryotes, is a well-studied calcium sensor. Apart from CaM, plants have a remarkable repertoire of putative Ca2+sensors that are not found in animals, including calmodulin-like (CML) proteins, calciumdependent protein kinases (CPKs), and calcineurin-B like proteins[12,16-18].Calmodulin contains four EF-hand motifs,whereas CML proteins contain variable numbers, ranging from one to six [12,16,19]. EF-hand motifs are the most prevalent structural motifs for Ca2+-binding [20]. Calmodulin regulates many cell processes, including gene regulation,protein synthesis, and ion homeostasis [12,16,21,22]. Although the Arabidopsis, tomato or rice genomes contain 50,52 and 51 CML proteins,respectively[12],the functional roles of only a few of them have been reported[16,19].For example,previous genetic studies showed that in Arabidopsis thaliana,CML24, CML9, and CML8 positively regulate defense against the bacterial pathogen Pseudomonas syringae[16,23,24].CML9 is also a negative regulator in abiotic stress response [25].Arabidopsis CML37 and CML42 play important roles in plant development and in responses to herbivores [16,26-28].Ectopic expression of a Glycine soja CML gene GsCML27 in Arabidopsis decreased plant tolerance to salt and osmotic stress but enhanced tolerance to bicarbonate stress during seed germination and early growth [29]. A recent study showed that overexpression of TaCML20, a calmodulin-like gene, enhanced water-soluble carbohydrate accumulation and yield in wheat [30]. However, there has been no report on CML response to pathogens in wheat.
To determine if and how CMLs are involved in response to R.cerealis infection in wheat we identified a CML gene named TaCML36. We made a detailed characterization of transcription profiles of this gene, investigated the protein subcellular localization,and dissected the defense function as well as the underlying mechanism. Our results revealed that TaCML36 positively contributes to the wheat immune response to R.cerealis by modulating expression of defense-associated genes, such as TaPIE1, Chitinase 1, PDF35, and PR17C. We identified a candidate gene for improving wheat resistance to sharp eyespot.
2. Methods and materials
2.1. Plants, fungal materials and treatments as well primers
Six wheat cultivars, including R. cerealis resistant cultivars CI 12633 and Shanhongmai, moderately-resistant cultivars Shangnong 0431 and Niavt 14, moderately-susceptible cultivar Yangmai 158,and susceptible cultivar Wenmai 6[8],were used to investigate TaCML36 transcription profiles. F9recombinant inbred lines (RIL) derived from the cross Shanhongmai × Wenmai 6 were provided by Prof. Jizeng Jia(CAAS). The highly virulent R. cerealis strain Rc207 was provided by Prof.Jinfen Yu(Shandong Agricultural University,Taishan).
Wheat seedlings were grown in a 14 h light (23 °C)/10 h darkness(12 °C)photoperiod.The second basal leaf sheath of each seedling was inoculated with small toothpick fragments harboring well-developed mycelia of R.cerealis or without the fungus (mock) following the protocol of Chen et al. [2]. RNA derived from three resistant and susceptible RIL at 4- and 10-days post inoculation(dpi)with R.cerealis and from the mock treatments were individually subjected to RNA-Seq-based transcriptomic analysis. Following protocols previously described by Zhang et al. [31] CI 12633 plants at the three-leaf stage were separately sprayed with solutions of phytohormones, including 0.05 mmol L-11-aminocyclopropane-1-carboxylic acid (ACC, an ethylene biosynthesis precursor),1.0 mmol L-1salicylic acid (SA), 0.1 mmol L-1methyl jasmonate (MeJA, a JA analog), and 0.2 mmol L-1abscisic acid(ABA).Leaf samples of plants treated with the hormones and non-sprayed controls were harvested at 0.5,1,3,6,12,and 24 h post-application, quickly frozen in liquid nitrogen, and stored at-80 °C prior to total RNA extraction.
All primers and their sequences are listed in Table S1.
2.2. RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol (Invitrogen, USA), and then subjected to RNase-free DNase I (TaKaRa, Japan) digestion and purification.
The purified RNA sample(2 μg)was reverse-transcribed to cDNA using the FastQuant RT Kit with gDNase(TaKaRa).
2.3. Isolation and sequence analysis of TaCML36
Transcriptomic analysis of RNA-sequence (RNA-Seq) data indicated that a wheat CML sequence was significantly induced after infection by R. cerealis. This gene sequence was cloned from the resistant wheat cultivar CI 12633 based on the above sequence TraesCS5D02G141900 downloaded from http://plants.ensembl.org/ and designated as TaCML36. The DNA sequence of TaCML36 was amplified in two rounds of nested PCR of DNA isolated from CI 12633 stem tissue.Primers for the first round of PCR were TaCML36-F1 (-580 to -561, 5′-AGTGGGCACAAAAGAGTTAG-3′) and TaCML36-R1 (+996 to+1016, 5′-CTATGCTGGACTACGGAACTA-3′), those for the second round were TaCML36-F2 (-381 to -360, 5′-TCGCATT TGATATGTATCTGGT-3′) and TaCML36-R2 (+598 to +617, 5′-AATGAACGAACAACGGTAGC-3′). The cDNA sequence of TaCML36 was amplified in two rounds of nested PCR of cDNA from CI 12633 stems 4 days post inoculation with R.cerealis strainRc207. Primers for the first round PCR were TaCML36-CF1 (-47 to -30 in the 5′-untranslated region (UTR),5′-CCTCCTGGTCCACTCTTT-3′) and TaCML36-CR1 (+696 to+716 in 3′-UTR, 5′-GCCTCTTCGTCGTGATTAGTT-3′); those for the second round were TaCML36-CF2 (-24 to -4 in 5′-UTR, 5′-GATACGTACGAGCCCAGGACA-3′) and TaCML36-CR2 (+605 to+628 in 3′-UTR,5′-TACTCACTAATAATGAACGAACAA-3′).
The deduced protein sequence of TaCML36 was obtained by means of the Pfam database (http://pfam.xfam.org/) and smart software (http://smart.embl-heidelberg.de/). Sequence alignment was performed by DNAMAN software.A neighborjoining phylogenetic tree was constructed using the MEGA V7.0 program with 1000 bootstrap replications.
2.4. Subcellular localization of TaCML36
The coding region of TaCML36 without the stop codon was amplified using gene-specific primers with Pst I and Bam HI restriction sites, and was subcloned in-frame to the Nterminus of the GFP sequence in the pJIT163:GFP vector. The resulting TaCML36-GFP fusion constructs p35S:TaCML36-GFP and GFP alone were introduced into wheat protoplasts via the PEG-mediated transfection method as previously described[32]. Transformed wheat protoplasts were incubated at 25 °C for 15 h.Following Qi et al.[33],GFP signals were observed and photographed using confocal laser scanning microscopy(Zeiss LSM 700, G?ttingen, Germany) with a Fluar 103/0.50 M27 objective lens and SP640 filter.
2.5. VIGS assays for defense function of TaCML36
To generate a BSMV:TaCML36 recombinant construct, a 285 bp sequence of TaCML36 (from 593 to 877 nucleotides in TaCML36 cDNA sequence) was sub-cloned in an antisense orientation into the Nhe I restriction site of the RNAγ part of barley stripe mosaic virus(BSMV)(Fig.S1).Following protocols previously described [34,35] the tripartite cDNA chains of BSMV:TaCML36 or the control BSMV:GFP virus genome were separately transcribed into RNAs, mixed, and used to inoculate CI 12633 plants at the three-leaf stage. The plants were grown in a 14 h light(23 °C)/10 h darkness(12 °C)photoperiod with ≥90% relative humidity for at least 48 h. At 10 days post virus transfection, fourth leaves of inoculated seedlings were collected to monitor BSMV infection. Transcription of BSMV coat protein gene CP detected by gene-specific primers(BSMVCPF and BSMV-CPR) was used to determine that BSMV had successfully infected the wheat plants.Real-time quantitative RT-PCR(RT-qPCR)with TaCML36 specific primers was used to examine TaCML36 transcript changes. The BSMV-infected CI 12633 plants were further inoculated with R. cerealis strain Rc207 mycelia 25 days post BSMV transfection and scored for fungal development 4 and 40 days later. Infection types (ITs)were categorized from 0 to 4 based on lesion size at the stem base[36],where IT 0 indicated no disease symptoms on the leaf sheath or stem;IT 1 described lesions appearing on the sheaths but not stems; IT 2, referred to lesion widths <50% of the infected stem perimeter; IT 3, referred to lesion widths >50%and <75%of the infected stem perimeter;and IT 4 indicated that the widths of lesions were >75%of the infected stem perimeter.A disease index (DI) = ∑ (di× li) × 100 / (L × di,max), where diindicated the infection type,liindicated the numbers of plants with each infection type, and L indicated the total number of plants.
2.6. Transcription of TaCML36 and defense-associated genes
RT-qPCR was used to investigate transcription of TaCML36 and five other wheat genes, viz. Chitinase 1 (GenBank accession number CA665185), PDF35 (GenBank accession number CA630387), PR17C (GenBank accession no. TA65181),TaPIE1 (GenBank accession number EF583940), and ACC oxidase 2 (ACO2, GenBank accession number AK332340). RTqPCR was performed using SYBR Green I Master Mix (Takara,Japan) in volumes of 20 μL on an ABI 7500 RT-PCR system(Applied Biosystems, USA). All RT-qPCR experiments for individual samples were repeated three times. The relative transcription levels of TaCML36 and five other genes were calculated using the 2-ΔΔCTmethod[37],where the wheat actin gene was used to normalize the amounts of cDNA among samples.
3. Results
3.1. Identification and sequencing of TaCML36
To identify wheat genes involved in the defense response to R.cerealis,RNA-seq was performed on the Shanhongmai × Wenmai 6 RILs at 4 and 10 dpi with R.cerealis.Comparative transcriptomic analysis showed that the TraesCS5D02G141900 sequence was highly homologous to the Aegilops tauschii CML36 gene. It was designated as TaCML36 and was up-regulated in the three resistant RILs.RNA-Seq data showed that TaCML36 transcription was 3.51-fold and 5.78-fold upregulated at 4 and 10 dpi relative to the non-inoculated mock treatment,respectively.
TaCML36 cDNA and genomic DNA sequences were cloned from resistant wheat cultivar CI 12633.The cDNA sequence of TaCML36 contained a 552 bp open reading frame, 316 bp 3′-UTR and a 106 bp 5′-UTR. The coding sequence of TaCML36 cloned from CI 12633 shared 100% identity with sequence TraesCS5D02G141900 located in wheat chromosome 5D.Comparison of the cDNA and genomic sequences showed that there was no intron in the TaCML36 genomic sequence.Because common wheat is hexaploid and contains A,B,D sets of genomes, three copies of TaCML36 were expected. After blasting the chromosome-based draft sequence of hexaploid wheat (https://urgi.versailles.inra.fr/) using the TaCML36 sequence on chromosome 5D, homeologs were identified on chromosomes 5AL and 5BL. The coding sequences of three copies on chromosomes 5AL, 5BL, and 5DL shared 98.91%identity(Fig.S2).The predicted TaCML36 protein derived from the gene sequence amplified from CI 12633 consisted of 183 amino acid residues with a predicted molecular weight of 19.07 kDa and an isoelectric point of 4.45. Protein sequence analysis showed that TaCML36 contained two highly conserved calcium-binding EF-hand domains(Fig.1).
A further blast-search of GenBank (https://blast.ncbi.nlm.nih.gov/) indicated that the sequence of TaCML36 hit Aegilops tauschii CML36 mRNA(AetCML36,GenBank No.XM_020302486.1, sharing 98.73% identity), and Hordeum vulgare ssp. vulgare mRNA for predicted protein (HvCML36, AK361300.1, sharing 93.12% identity). We constructed a phylogenetic tree, including TaCML36, AetCML36, HvCML36, TaCML20, GsCML27,GmCAM4, and 19 known-function Arabidopsis CML proteins(Fig. S3). TaCML36 and other CML36 proteins clustered into a clade (Fig. S3). TaCML36 shared the highest amino acid sequence identity (98.36%) with AetCML36 and shared 33.77% identity with Arabidopsis CML36 (AtCML36). Both AetCML36 and TaCML36 contained two EF-hand domains,whereas AtCML36 possessed four. Further amino acid sequence alignment indicated that the two EF-hand motifs of TaCML36 matched the last two (nearer the C-terminal sequence) of AtCML36 where the EF-hand motifs shared 100%sequence identity.
3.2. TaCML36 transcriptional profiles point to a defense response to R. cerealis infection
RT-qPCR results showed transcription levels of TaCML36 at 10 dpi with R. cerealis were significantly higher in resistant wheat CI 12633, Shanhongmai, Niavt 14, and Shannong 0431 than in susceptible cultivars Yangmai 158 and Wenmai 6(Fig.2-a). TaCML36 transcription in CI 12633 was higher at both 4 dpi and 10 dpi, whereas that in Wenmai 6 had declined (Fig. 2-b).Tissue expression analysis showed that transcription levels of TaCML36 were highest in sheaths, followed by spikes and lowest in stems, whereas the largest transcription increase in CI 12633 compared to Wenmai 6 occurred in the stems where disease symptoms appear (a 9.33-fold change) (Fig. 2-c). These results suggested that TaCML36 might participate in the wheat immune response to sharp eyespot.
3.3. TaCML36 is responsive to external ethylene, MeJA, ABA and SA stimuli
We investigated the transcription profiles of TaCML36 in wheat leaves treated with ACC (an ethylene biosynthesis precursor),MeJA,SA and ABA,respectively,to determine how TaCML36 responds to exogenous applications of hormones(Fig. 3). The fastest and highest induction effects occurred with ACC treatment, where expression was induced from 0.5 to 24 h post-treatment (hpt), rapidly increased to 8.60-fold at 0.5 hpt,reached a peak at 1 hpt(about 10.89-fold that of nontreatment), then decreased to 4.67-fold higher than the nontreated control (N) from 3 to 24 hpt (Fig. 3-a). After treatment with MeJA,TaCML36 transcription induction reached a peak at 0.5 hpt(about 4.45-fold the control),decreased at 3 hpt(about 0.58-fold below that of non-treatment), then increased to an average level of 3.08-fold during 6-24 hpt(Fig.3-b).Upon ABA treatment, the transcription level of TaCML36 was induced from 0.5 to 24 hpt except for 12 hpt when the level had decreased to 0.53-fold above that of non-treatment (Fig. 3-c).After SA treatment, the TaCML36 transcriptional level decreased to 0.55-fold at 0.5 hpt and then increased slowly during 1-12 hpt,reaching a peak at 12 hpt that was 21.19-fold above that of the non-treatment (Fig. 3-d). These results suggested that TaCML36 transcription was induced by exogenous ethylene,JA,and ABA stimuli,but response to external SA treatment was more complex.
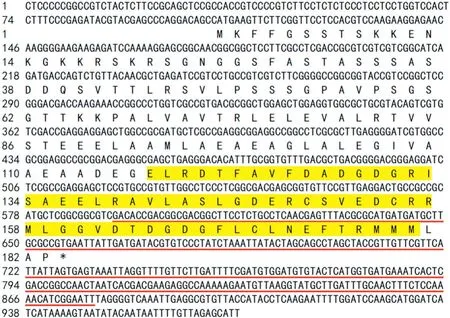
Fig.1-Nucleotide sequence of TaCML36 and its deduced amino acid sequence.Yellow highlighting shows the EF-hand domains.The 285 bp TaCML36 fragment(from 593 to 877 nt in the TaCML36 cDNA sequence)underlined in red was sub-cloned in an antisense orientation into the Nhe I restriction site of the RNAγ part of BSMV.
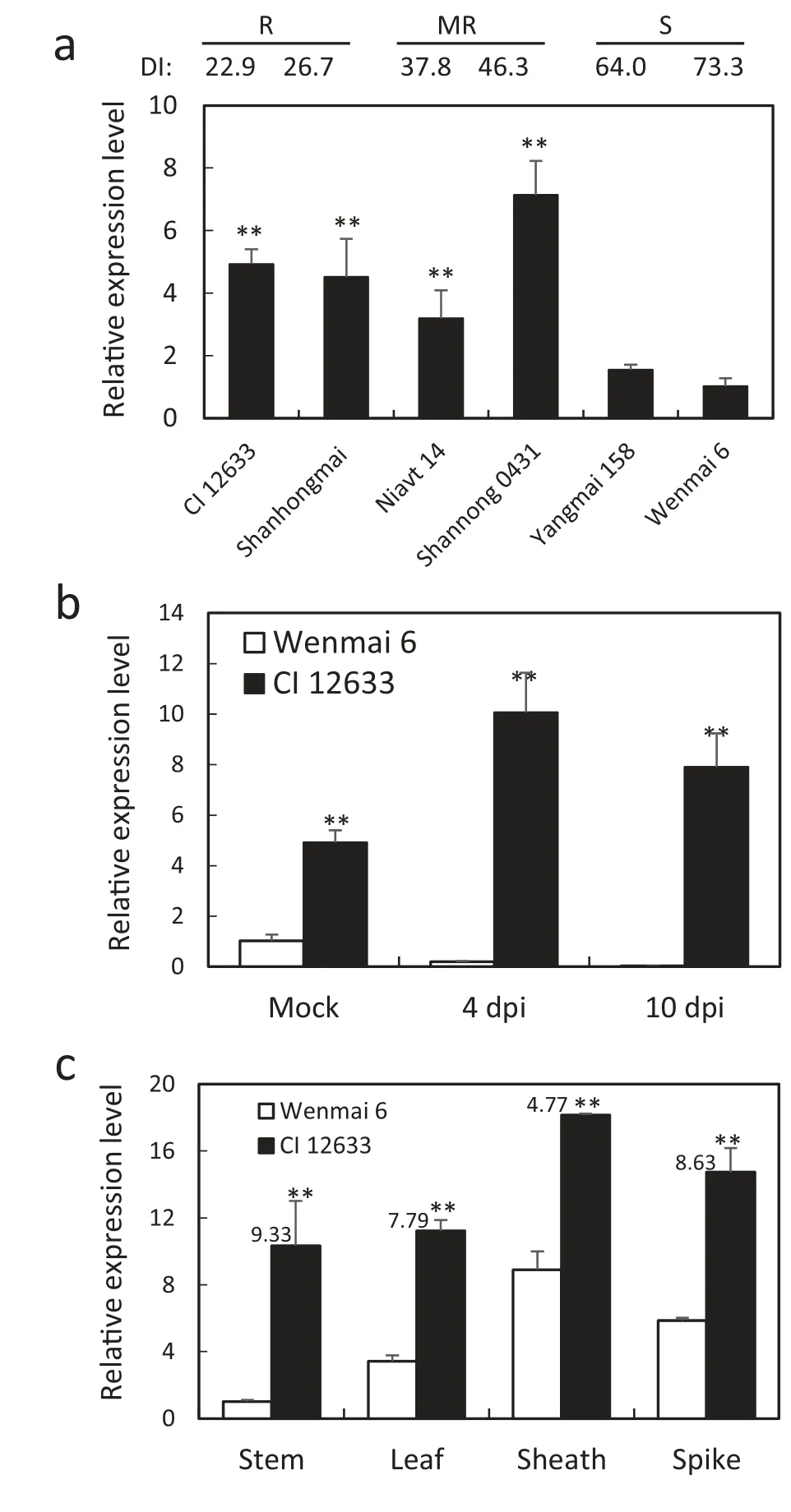
Fig.2-Transcription patterns of TaCML36 determined by RTqPCR in wheat following inoculation with R. cerealis strain Rc207.(a) Relative transcriptional levels of TaCML36 resistant,moderately-resistant,moderately-susceptible and susceptible wheat cultivars were measured 10 dpi.Transcriptional levels are relative to that of Wenmai 6.DI indicates disease index.(b)Relative transcription levels of TaCML36 in resistant cultivar CI 12633 and susceptible cultivar Wenmai 6 at 4 and 10 dpi or in mock-treatments with no pathogen. (c)Transcription of TaCML36 in stems,leaves,leaf sheaths,and spikes of CI 12633 and Wenmai 6 at 40 dpi. Significant differences between the cultivars were based on results from three replications (Student's t-tests:**P <0.01).Error bars indicate standard errors.
3.4. TaCML36 protein localizes the nucleus and cytoplasm
To investigate the subcellular localization of TaCML36 the coding region of TaCML36 was fused in-frame with GFP.TaCML36-GFP and the GFP control were then transiently expressed separately in wheat protoplasts. As shown in Fig.4 TaCML36-GFP was present in both the cytoplasm and nucleus in wheat protoplasts whereas it was localized to the entire cell in the GFP control(Fig.4).
3.5. Silencing of TaCML36 in wheat reduced resistance sharp eyespot
To assess defense function of TaCML36, a cDNA fragment of TaCML36 was used to construct a BSMV-based VIGS vector and deployed to knock-down TaCML36 in cultivar CI 12633. BSMV symptoms became evident in both BSMV:GFP- and BSMV:TaCML36-inoculated plants at 10 dpi with the virus (Fig. 5-a)when transcripts of the BSMV cp gene were readily detected(Fig. 5-b), indicating that BSMV infection was successful.TaCML36 transcription levels were substantially reduced in BSMV:TaCML36-infected wheat plants, indicating that TaCML36 had been silenced (Fig. 5-c). The BSMV:GFP- and BSMV:TaCML36-infected CI 12633 plants were inoculated with R.cerealis to assess the defense role of TaCML36.
Sharp eyespot symptoms (brown lesions) appeared on inoculated leaf sheaths and stems of BSMV:TaCML36-infected CI 12633 at 4 dpi, but was absent on stems of inoculated BSMV:GFP-treated plants (Fig. 5-d). Using R. cerealis RcActin transcriptional levels as an indicator of the fungal biomass,relative R. cerealis abundance in the BSMV:TaCML36-infected CI 12633 plants was significantly higher than in BSMV:GFPtreated plants (Fig. 5-e). This indicated that down-regulation of TaCML36 in CI 12633 led to higher susceptibility. More intense symptoms on the stems of BSMV:TaCML36-infected CI 12633 plants were present at 40 dpi (average DI: 42.72 ±14.20)compared with those on stems of BSMV:GFP-treated CI 12633 plants (average DI: 30.00 ± 10.30) (P <0.01) (Fig. 5-f).Thus knock-down of TaCML36 in CI 12633 impaired its resistance to sharp eyespot suggesting that TaCML36 positively regulated the immune response.
3.6.Silencing of TaCML36 downregulates a subset of defenserelated genes
To explore molecular mechanism underlying the defense role of TaCML36, we investigated by RT-qPCR whether or not silencing of TaCML36 would affect the transcript levels of wheat defense response genes. The tested genes included Chitinase 1, belonging to the pathogenesis-related-3 (PR-3)family [2], PDF35, belonging to the PR-12 family, ethylene response factor gene TaPIE1[38],and ACC oxidase gene ACO2.The transcription levels of Chitinase 1, PDF35, PR17C, TaPIE1,and ACO2, were significantly decreased in TaCML36-silenced plants relative to BSMV:GFP-infected controls (Fig. 6). These data suggest that TaCML36 positively regulates transcription of these defense-associated genes.
4. Discussion
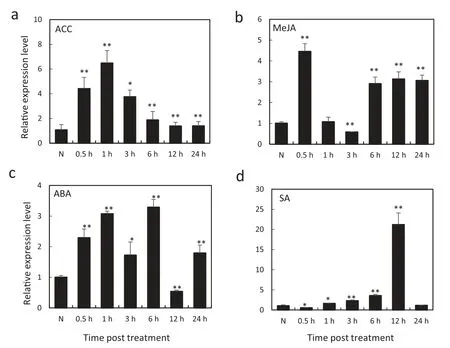
Fig.3-Expression patterns of TaCML36 in wheat after treatment with exogenous hormones ACC(a),MeJA(b),ABA(c),and SA(d)for 0.5,1,3,6,12,and 24 h.Relative expression of TaCML36 is shown as fold change in transcription over the non-treatment control(N).Three biological replicates for each time point were averaged,with the standard error indicated.Asterisks indicate statistically significant differences from the control based on Student's t-tests(*P <0.05,**P <0.01).
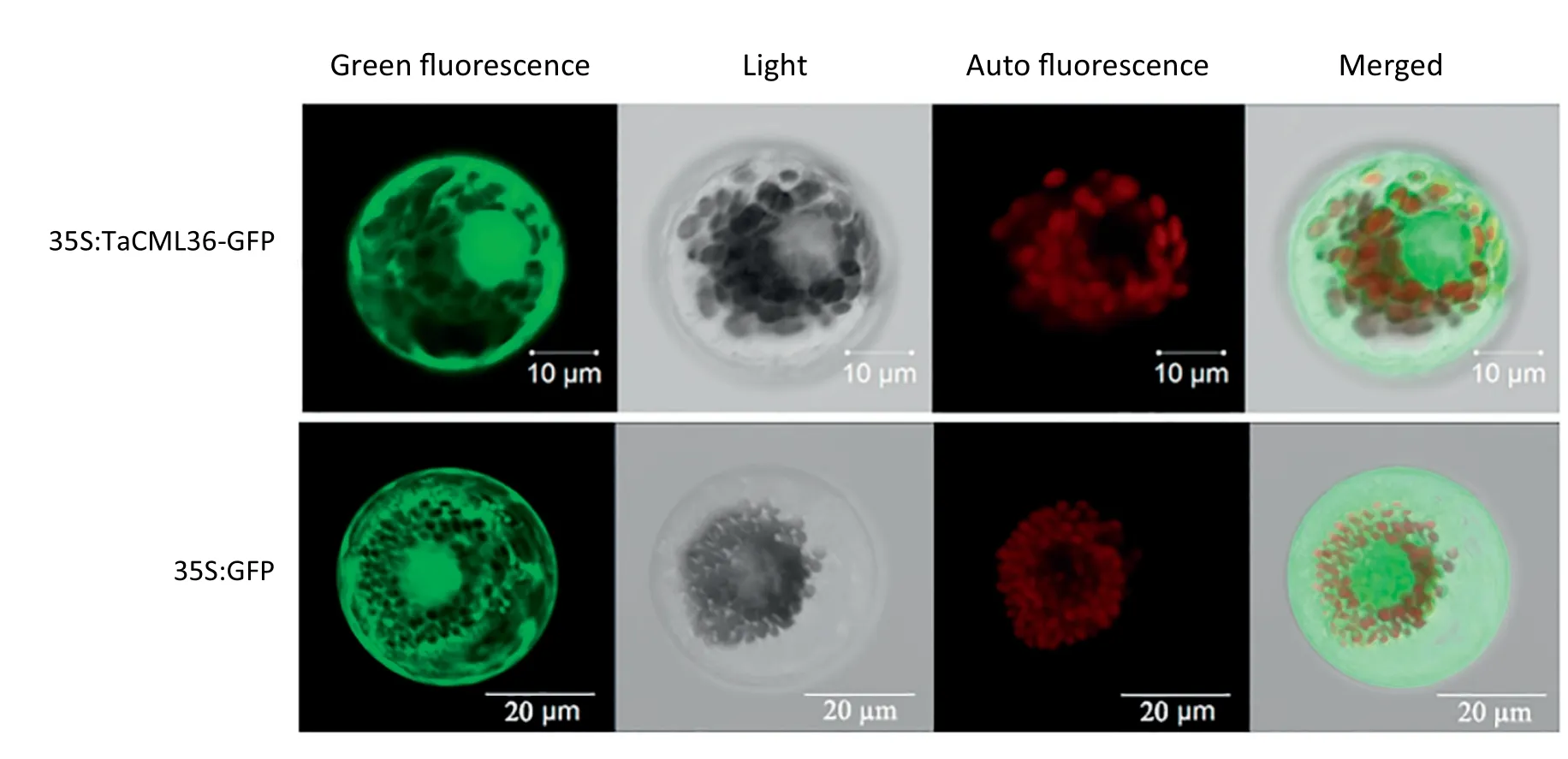
Fig.4- Subcellular localization of TaCML36 in wheat mesophyll protoplasts. 35S:TaCML36-GFP and 35S:GFP constructs were individually introduced into wheat mesophyll protoplasts.Transient expression of green-fluorescence proteins(GFP)was observed and photographed under a confocal laser scanning microscope(Zeiss LSM 700, G?ttingen, Germany). Autofluorescence of the wheat chloroplasts was also observed and photographed.
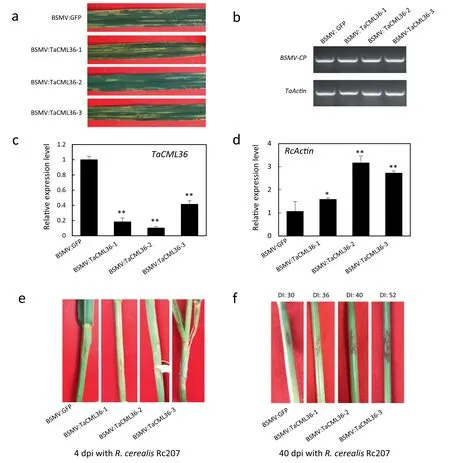
Fig.5-Molecular characterization and R.cerealis responses to TaCML36-silencing in CI 12633 plants.(a)Mild chlorotic mosaic symptoms were observed on leaves at 10 days dpi when separately transfected with BSMV:GFP or BSMV:TaCML36.(b) RTqPCR analysis of transcription levels of the BSMV coat protein gene in leaves of BSMV:GFP- and BSMV:TaCML36-infected CI 12633 plants at 10 days post virus inoculation.(c)RT-qPCR analysis of transcription levels of the TaCML36 gene in leaves of BSMV:GFP-and BSMV:TaCML36-infected CI 12633 plants at 10 days post virus inoculation.Error bars were calculated based on three replicates.Statistically significant differences between BSMV:TaCML36-and BSMV:GFP-inoculated plants were determined by Student's t-tests(**P <0.01).(d)RT-qPCR analysis of the R.cerealis actin(RcActin)gene in stems of TaCML36-silenced and control CI 12633 plants at 4 days following R.cerealis inoculation represented the biomass of R.cerealis.Error bars were based on three replicates.Statistically significant differences between BSMV:TaCML36-and BSMV:GFP-inoculated plants were determined based by Student's t-tests(*P <0.05;**,P <0.01).(e)Sharp eyespot symptoms of BSMV:GFP-and BSMV:TaCML36-infected CI12633 plants for 4 dpi with R. cerealis.(f)Sharp eyespot symptoms of BSMV:GFP- and BSMV:TaCML36-infected CI 12633 plants at 40 dpi with R.cerealis.DI,sharp eyespot disease index.
Several fungal diseases, including sharp eyespot, impact wheat production worldwide. Isolation and functional characterization of important defense-related genes in wheat are vital for development of wheat varieties with resistance to fungal diseases. Previous studies showed that Arabidopsis CML24, CML8, and CML9, acting as calcium sensors, participate in the innate immune response to the bacterial pathogen Pseudomonas syringae[16,23,24].Over-expression of GmCaM4,a soybean CML, enhanced resistance of soybean to the oomycete Phytophthora sojae and necrotrophic fungal pathogens Alternaria tenuissima and Phomopsis longicolla [39]. Yet,little was known about the role of CMLs in immune responses in wheat.Here,we identified and proved that wheat CML gene TaCML36 participated in an immune response to R.cerealis.
The transcription level of TaCML36 was not only significantly elevated after infection by R. cerealis, but was also higher in sharp eyespot-resistant wheat cultivars than in susceptible wheat cultivars. In previous studies, Arabidopsis CML8 and CML9 were induced following infection by P.syringae and shown to positively regulate an innate immune response to infection by P.syringae[16,24].Loss of function of CML37 led to a better feeding performance of larvae,suggesting that CML37 is a positive regulator in defense response to herbivores [28].Also,the expression of TaCML20 is consistent with its role in drought response in wheat plants [30]. In the current research, knock-down of TaCML36 in resistant wheat cultivar CI 12633 by VIGS permitted growth of R.cerealis in the silenced wheat plants and in turn enhanced host susceptibility to infection by the pathogen. These results indicated that TaCML36 positively regulated an immune response to R.cerealis. To our knowledge, this is the first report uncovering a regulatory role of a monocotyledonous CML protein in plant immune response. Undoubtedly, this study broadens our understanding of biological function of CMLs in plant species.
Accumulating evidence indicates that CMLs modulate transcription of defense-associated genes,ultimately contributing to plant defense against infection by pathogens[16,23,24,39]. For instance, Arabidopsis CML8 and CML9 upregulate the expression of SA-dependent PR1 [16,24].GmCaM4 overexpression activates PR4 and increases transcription levels of PR1a, PR2, and PR3, whereas expression of these genes was repressed in GmCaM4-silenced plants[39].In this study, expression of Chitinase 1 (PR-3), PDF35 (PR-12),PR17C, TaPIE1, and ACO2 was lower in TaCML36-silenced wheat plants than in control plants, suggesting that TaCML36 positively regulates transcription of these genes.Chitinases and defensins (like PDF35 and RsAFP2) directly contribute to resistance to infection and growth of R. cerealis[40-44]. TaPIE1 was shown to participate in the ethylene signaling pathway, and to upregulate expression of late defense-related genes, such as defensin and Chitinase, in response to R. cerealis [38]. Comparing our results with those in previous studies [16,24,39], we found that TaCML36,GmCaM4, and Arabidopsis CML8 and CML9 possibly participate in plant immune responses to different life-style pathogens through modulating transcription levels of different subsets of PR.

Fig.6-Transcription levels of the TaCML36,TaACO2,TaPIE1,and PR(Chitinase1,PDF35,and PR17C)genes in BSMV:TaCML36-and BSMV:GFP-infected CI 12633 plants 4 dpi with R.cerealis.Statistically significant differences between BSMV:TaCML36-and BSMV:GFP-infected CI 12633 plants were determined by Student's t-tests(*P <0.05;**P <0.01).
Phytohormones,including SA,JA,ethylene,and ABA,have been implicated in plant response to both biotic and abiotic stresses. Some CMLs are involved in the SA, JA, and ABA signaling pathways during response to both biotic and abiotic stress.For example,Arabidopsis CML8 and CML9 are induced by SA treatment and increase SA-dependent PR1 transcription in the immune response to P. syringae [16,24]. In Arabidopsis,CML37 expression is induced by both JA and ABA. CML37 is a positive regulator of JA-mediated herbivore defense and ABAdependent drought responses [28,45]. The current investigation showed that although TaCML36 was responsive to external ethylene, JA, ABA, and SA treatments, the transcription induction of TaCML36 by ethylene was the most rapid and largest early response (0.5 and 1 hpt). Transcription of ACO2 was repressed in TaCML36-silenced wheat plants compared non-silenced controls, suggesting that TaCML36 positively regulates ACO2 transcription. ACO2 is a key enzyme in ethylene biosynthesis and can convert ACC to ethylene [46].Accordingly, ethylene level was lower in TaCML36-silenced wheat plants than in control plants(data not shown).Previous studies indicated that wheat Chitinase 1, Defensin(PDF35), and TaPIE1 were induced by external ethylene stimuli and are thus located downstream in the ethylene signaling pathway[2,38].Collectively, our results suggest that elevated expression of TaCML36 following infection might up-regulate transcription of defense-associated genes in the ethylene signaling pathway, resulting in enhanced resistance to sharp eyespot. To our knowledge this is the first report of CML involvement in the ethylene signaling pathway during a plant immune response.
Gene TaCML36 was first identified as matching the genomic sequence of TraesCS5D02G141900. The cDNA and gDNA sequences cloned from resistant cultivar CI 12633 were identical to those in Chinese Spring. The TaCML36 ORF sequence shared 97.83% identity with the its ortholog on chromosome 5A and 97.64% identity with the copy on chromosome 5B. These data imply that TaCML36 is highly conserved in wheat. No intron was present in the TaCML36 genomic sequence, consistent with reports that the majority of CMLs were intronless [47]. Protein sequence analysis indicated that TaCML36 contained two EF-hand domains and like most CML lacked particular localization signatures[30,48].Phylogenetic analysis showed that TaCML36 clustered with CML36 proteins,whose roles remain unknown except for TaCML36, which shares 33.77% identity with Arabidopsis CML36 (AtCML36), where AtCML36 possesses 4 calciumbinding EF-hand domains. Further EF-hand domain comparisons revealed that the two EF-hand domains in TaCML36 are identical to the last two EF-hand domains of AtCML36, but differ substantially from EF-hand domains of Arabidopsis CML24, CML8, and CML9. TaCML36 lacks the two EF-hand domains nearest the N-terminal sequence of AtCML36.Additionally, subcellular localization assays showed that TaCML36 localized in both the cytoplasm and nucleus.Several CML proteins, such as GsCML27, TaCML20, and GmCaM4,were likewise shown to be located in both the cytoplasm and nucleus[29,30,39].The nucleus-cytoplasmic distribution characteristic of these CMLs is possibly associated with their roles in activating proteins from the nucleus and cytoplasm[30,48].
5. Conclusions
The wheat CML gene TaCML36 was identified from RNA-Seq data and its sequence was cloned from the sharp eyespotresistant wheat cultivar CI 12633. TaCML36 transcription was significantly elevated by both R.cerealis infection and external ethylene treatment. TaCML36 positively contributed to the wheat innate immune response to R.cerealis infection through upregulation of transcription of a subset of defenseassociated genes mainly in the ethylene signaling pathway.Thus,TaCML36 is a candidate to improve resistance of wheat to sharp eyespot. This study also provides insights into the roles of CML proteins in plants.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2019.02.001.
Acknowledgments
The authors are very grateful to Prof. Jizeng Jia (Institute of Crop Sciences, CAAS) for providing the RIL population, Prof.Jinfeng Yu (Shandong Agricultural University, Tai’an, Shandong) for providing R. cerealis strain Rc207, and Dr. Hua Qin and Prof. Rongfeng Huang (Biotechnology Research Institute,CAAS)for examining ethylene contents in wheat leaves.This work was funded by the National “Key Sci-Tech” Project(2016ZX08002-001-004).
- The Crop Journal的其它文章
- OstMAPKKK5,a truncated mitogen-activated protein kinase kinase kinase 5,positively regulates plant height and yield in rice
- Mapping QTL affecting the vertical distribution and seed set of soybean[Glycine max(L.) Merr.]pods
- Identifying key traits in high-yielding rice cultivars for adaptability to both temperate and tropical environments
- Deep genotyping of the gene GmSNAP facilitates pyramiding resistance to cyst nematode in soybean
- Molecular mapping and candidate gene analysis of the semi-dominant gene Vestigial glume1 in maize
- Draft genome sequence of a less-known wild Vigna: Beach pea (V. marina cv. ANBp-14-03)