Expanding luminal epitheliums as cells of origin for prostate cancer
Despite tremendous progress made in human prostate biology over the past few decades,a full picture of prostate lineage hierarchy and its connection to cancer initiation remain to be delineated.Two recent studies published in
Nat Genet.
[1]and inScience
[2]have profiled subpopulations of prostate cells at the single-cell level.Complementary analyses of data from the studies demonstrate self-renewal and differentiation capacities of different luminal epithelial cells,which can serve as cells of origin for prostate cancer.The system of lineage hierarchy controls cell differentiation and tissue formation from stem and progenitor cells,which may initiate cancer development if the process goes awry[3].Therefore,it is of great interest to identify cells with stemness or progenitor properties and delineate their lineage hierarchy in order to pinpoint the cells of origin for cancer when cellular genetics are altered[4].
The prostate epithelium is composed of luminal,basal and a rare population of neuroendocrine(NE)cells[5].Previous studies have identified progenitor cells in both luminal and basal populations,which can serve as cells of origin for prostate cancer[6,7].However,these studies were all carried out in engineered mouse models,where the initiation of murine prostate cancer might be very different from those in humans under pathophysiological conditions.In addition,previous studies mainly used bulk cells of luminal or basal subtypes,which hindered the identification of relatively rare stem or progenitor cells due to the heterogenous nature of cell populations.
The advent of single-cell profiling has facilitated unprecedented stratification of cells within tissues,which can help to identify rare populations of cells[8].Recently,in two independent studies,Guo et al.[1]and Karthaus et al.[2]used droplet-based single-cell RNA sequencing(scRNAseq)to profile prostate cells isolated from mice,and each identified three clusters of luminal epithelial cells.Based on the markers used for clustering,the Luminal-A and Luminal-B cells identified by Guo et al.[1]were largely overlapping the L1 population identified by Karthaus et al.[2],as they were all Nkx3.1and localized in the distal regions of murine prostate.The L2 population of cells identified by Karthaus et al.[2]mainly localized in the proximal prostate,which was also revealed in the other study and was designated as Luminal-C cells.The L2/Luminal-C cells are positive for stemness genesTacstd2
andPsca
,and they are apparently the same progenitor cells identified as Sca-1/Nkx3.1cells from a previous study[9].While Karthaus et al.[2]only analyzed cells from the anterior prostate(AP),Guo et al.[1]profiled all three prostate lobes:AP,ventral prostate(VP)and dorsal-lateral prostate(DLP),which revealed lobe-specific distributions of the luminal clusters.Notably,Guo et al.[1]also noticed the existence of some Luminal-C cells at the invagination tips of prostate distal region,for which they separately designated these cells as Prox-Luminal-C and Dist-Luminal-C cells(Fig.1).Guo et al.[1]then continued to functionally characterize different clusters of prostate luminal cells.Markovchain entropy analysis showed that both Prox-and Dist-Luminal-C cells had higher stemness potency scores than Tacstd2luminal cells.Indeed,Tacstd2Prox-and Dist-Luminal-C cells isolated from mice formed organoids at efficiencies three and six times higher,respectively,than Tacstd2luminal cells(Fig.1).Renal capsule grafting also demonstrated a much higher capacity of Luminal-C cells in the regeneration of prostate glands than Tacstd2cells.Remarkably,Luminal-C cells generated both luminal and basal cells in these two assays,and Tacstd2cells still resided at the invagination tips of regenerated prostate glands,which also had newly produced Tacstd2Luminal-A and Luminal-B daughter cells(Fig.1).Additionally,lineagetracing experiment showed consistent localization of Dist-Luminal-C cells at the invagination tips of prostate distal regions.Similarly,in a prostate regression-regeneration assay,Dist-Luminal-C cells not only sustained Tacstd2cells,but also generated Tacstd2luminal cells[1].
Karthaus et al.[2]also showed that L2 cells isolated from intact mice(no castration)had a higher organoidforming efficiency than L1 cells;L2 cells were less sensitive to castration than L1 cells in the organoid formation assay,consistent with a higher stemness potency of L2/Luminal-C cells.In a lineage-tracing experiment with luminal-specific Krt8 Credriver,the authors showed that luminal cells reconstituted prostate glands at their original proximal or distal regions,suggesting each population contains its own progenitor cells.Both L1 and L2 cells regenerated Ck5basal cells in organoids and reconstituted prostate glands,consistent with the other study.In addition,Karthaus et al.[2]demonstrated that luminal cell growth in organoid culture was supported by growth factors derived from mesenchymal cells in a paracrine manner(Fig.1).They identified several mesenchyme-secreted growth factors,whose expressions were changed during castration/regeneration cycle,as well as their receptors on luminal cells[2].These data highlight the importance of complex stromal systems in regulating prostate epithelial cells,and may explain certain phenotypical differencesin vivo
andin vitro
.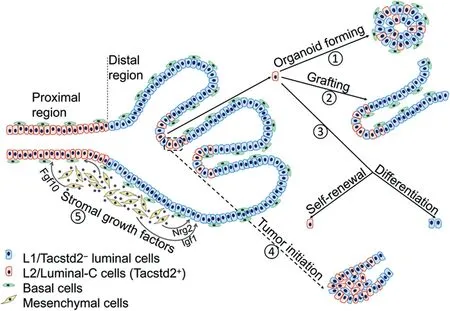
Figure 1 Tacstd2+L2/Luminal-C cells have stemness properties and can serve as cells of origin for prostate cancer.Tacstd2+Luminal-C cells are localized to the invagination tips of prostate distal regions,whereas in the proximal region,they form a continuous luminal layer.①Tacstd2+Luminal-C cells isolated from both proximal and distal regions can form organoids that have both luminal and basal epithelial cells.②Prostate gland reconstructed from Luminal-C cells by grafting contains both luminal and basal cell markers,and Tacstd2+Luminal-C cells are confined to the invagination tips of the regenerated gland.③Tacstd2+Luminal-C cells isolated from prostate distal regions have the capacity of self-renewal and differentiation into Tacstd2-luminal cells.④Dist-Luminal-C cells initiate prostate cancer after lineage-specific deletion of the tumor suppressor gene Pten(dashed rectangle).⑤Growth factors secreted by mesenchymal cells support luminal cell growth.Fgf10,fibroblast growth factor 10;Igf1,insulinlike growth factor 1;Nrg2,neuregulin 2.
Given the self-renewal and differentiation capacities of Luminal-C cells,Guo et al.[1]further sought to elucidate implications of these cells in prostate cancer development.Luminal-C lineage-specific deletion of tumor suppressor genePten
initiated prostate hyperplasia,which would progress to prostatic intraepithelial neoplasia(PIN)after longer time.Compared to Luminal-C cells from prostate proximal regions,Dist-Luminal-C cells showed higher efficiency of prostate neoplasia initiation,in line with their more efficient organoid-forming capability than Prox-Luminal-C cells.Luminal-C cells also showed faster tumor initiation than Tacstd2Luminal-A and Luminal-B cells.This is likely caused by dedifferentiation of Luminal-A and Luminal-B cells into Luminal-C cells before initiating cancer,further emphasizing Luminal-C cells as progenitors for prostate cancer,a conclusion similarly drawn by Karthaus et al.from their study[2].Finally,both studies applied scRNA-seq to profile human prostate tissues,which showed not only similarities but also significant differences when compared to murine prostate.For example,the seven clusters of cells identified by Guo et al.[1]distributed in all three human prostate zones,namely central,peripheral and transition zones.On the other hand,Karthaus et al.[2]identified two luminal clusters mostly featuring the L1 and L2 cells from mouse prostate,but there were cells expressing both L1 and L2 markers,which were suggested to be possible bipotent progenitor cells.These findings are consistent with the notion that human and mouse prostates have significant anatomical differences.Nevertheless,Guo et al.[1]showed the existence of TACSTD2Luminal-C cells in normal human prostate,which also localized at the invagination tips.Analyses of prostate cancer datasets revealed that TACSTD2 expression is significantly higher in prostate tumors than in normal prostate,and its expression levels are correlated with Gleason scores of the prostate tumors[1].These studies confirm the postulations that TACSTD2L2-like or Luminal-C epithelial cells can serve as cellular origins for prostate cancer.
In the organoid formation and renal capsule grafting assays,Luminal-C cells generated both luminal and basal cells,whereas in the lineage-tracing and regressionregeneration assaysin vivo
,all lineage-marked cells were positive for luminal marker CK8,but negative for basal makers(Fig.1),indicating that Luminal-C cells responded differentlyin vitro
andin vivo
.It would be useful in the future to define those factors that determine the differentiation commitment of Luminal-C cells,which may explain the predominantly luminal phenotypes of prostate tumors[6].It is worth noting that a study on airway epithelial cell dedifferentiation[10]indicated that ablation of stem cells provides an intrinsic stimulus for luminal cell to regain pluripotency,which may give a hint as to how luminal prostate cells produce basal cellsex vivo
.Author contributions
Study design
:Gonghong Wei.Data acquisition
:Yuexi Gu,Gonghong Wei.Data analysis
:Yuexi Gu,Gonghong Wei.Drafting of manuscript
:Yuexi Gu,Gonghong Wei.Critical revision of the manuscript
:Yuexi Gu,Gonghong Wei.Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by Fudan University Recruitment Grant and the the Jane and Aatos Erkko Foundation.
Yuexi Gu
Fudan University Shanghai Cancer Center,Department of Biochemistry and Molecular Biology,School of Basic Medical Sciences,Shanghai Medical College of Fudan University,Shanghai,China
Gong-Hong Wei
Fudan University Shanghai Cancer Center,Department of Biochemistry and Molecular Biology,School of Basic Medical Sciences,Shanghai Medical College of Fudan University,Shanghai,China
Biocenter Oulu and Faculty of Biochemistry and Molecular Medicine,University of Oulu,Oulu,Finland
*Corresponding author.
E-mail addresses:
gu-yuexi@hotmail.com(Y.Gu),gonghong_wei@fudan.edu.cn(G.-H.Wei)4 September 2020
 Asian Journal of Urology2021年2期
Asian Journal of Urology2021年2期
- Asian Journal of Urology的其它文章
- Super-mini percutaneous nephrolithotomy
- Are we progressing in prostate cancer management?
- Testicular germ cell tumors
- Multimodal therapy in oligometastatic prostate cancer:A glimpse into the future?
- Bilateral ureteral reconstruction using appendicular interposition combined with Wallace anastomosis following stenosis after radical cystectomy and ileal neobladder construction
- Cystic nephroma:A bosniak III benign tumor in the kidney
