Phylogenetic Analysis of Legionella Strains and ldentification of Serogroups by Lipopolysaccharide-and O-antigen-based PCR Assay*
LI Yi Nan , REN Hong Yu , ZHAO Na , WANG Yan Qing,2 , LI Dai , and QIN Tian,3,#
Legionella,a genus of pathogenic Gram-negative bacteria,is widely present in natural water sources and artificial water systems.A total of 65 species and> 70 serogroups ofLegionellahave been characterized[1].Lipopolysaccharide (LPS),the main component of the outer membrane ofLegionella,is not only associated with toxicity,but also provides the basis for classification in serotyping[2].LPS consists of lipid A,core polysaccharide,and Oantigen.O-antigens are glycopolymers expressed on the cell surface of Gram-negative bacteria.Variability in the O-antigen structure constitutes the basis for the establishment of serotyping.Based on variations in the O-antigen,Legionellacan be divided into 15 serogroups[3]. Approximately 84% ofLegionellainfections are caused byL.pneumophilaserogroup 1(LP1)[4],and Legionnaires’ disease caused by non-L.pneumophilastrains ofLegionellaaccounts for approximately 5%-10% of all cases[5].
LPS comprises 0-1.6% of the dry weight ofL.pneumophila.LPS has been shown to stimulate morphological changes and alterations in gene expression in almost all host cells,leading to the uncontrolled expression of host cytokines,severe infection,and septic shock[6].Gene clusters encoding enzymes for the synthesis ofLegionellaLPS provide model candidates for studying molecular evolution at the DNA level.TheLegionellaO-antigen contains a number of isomers and derivatives of the monosaccharide pseudaminic acid,such as legionaminic acid and 4-epilegionaminic acid[7].This monosaccharide is rarely present in the O-antigen of bacteria,thus accounting for the unique O-antigen structure ofLegionella.
In the present study,variousLegionellaserogroups were identified by PCR,and comparisons were made among the O-antigen-specific genes.The PCR method has been tested previously for its specificity and sensitivity[8,9],and LPS composition may be a determinant of serogroup specificity,as defined by the immunofluorescence-based serotyping schema forL.pneumophilaand otherLegionellaspecies[2].
Whole genome sequences from 97 strains ofLegionellawere analyzed in the present study.Complete details of these strains,including the strain name,source,place of isolation,serotype,and time of isolation,are provided in Supplementary Table S1 available in www.besjournal.com. The data generated in this Whole Genome Shotgun project have been deposited in the National Center for Biotechnology Information (NCBI) under the BioProjectID PRJNA281151 with accession numbers LBAW00000000,LBHK00000000,LBAX00000000,LUCB00000000,LCUA00000000,LBAY00000000,LAVP00000000,and LBMS00000000.
The LPS synthesis gene sequences of allLegionellastrains were downloaded in NCBI and multiple annotated databases (including NR,SwissProt,KEGG,COG,TCDB,go,PHI,VFDB,ARDB,Secretory-protein,T3SS,and CAZy).The LPS synthesis gene structure of each serogroup ofLegionellawas found to be similar.Among the LPS core genes of 15 serotypes (including serogroups LP1-14 and an unknown),the genomes of serotypes LP1,LP2,LP3,LP4,LP5,LP7,LP8,LP9,LP10,and LP13 contained 19 core genes.LP11 contained fewer LPS genes than LP1-LP5,LP7-LP10,and LP13,and lacked the genes lpg0837_YP_094872.1,lpg0838_YP_094873.1,lpg0920_YP_094954.1,lpg0749_YP_094785.1,lpg2630_YP_096635.1,lpxD_lpg2944_YP_096937.1,lpg2695_YP_096700.1,lpxB_lpg2945_YP_096938.1,lpg0748_YP_094784.1,and lpg2799_YP_096794.1 (Supplementary Figure S1,available in www.besjournal.com).
Core-pan analysis of all serogroups (LP1-14 and the unknown) was then performed.Numbers of LPS core and pan genes,and the core-pan differences between each serogroup ofLegionella,are shown in Supplementary Table S2 available in www.besjournal.com.For the pan genes of LPS,the following differences were observed:Among the 15 serotypes,LP1 and the unknown contained the lpg0777_YP_094813.1 gene (an O-acetyltransferase),and unlike the otherLegionellatypes,LP4,LP5,LP8,LP9,LP11,and LP13 lacked the lpg0782_YP_094818.1 gene (encoding a putative Oacetyltransferase).Among the core genes for LPS synthesis,LP14 uniquely contained a gene,alpg2600_YP_ 096605.1,whereas LP11 contained fewer LPS pan genes compared with other serogroups ofLegionella(Supplementary Figure S2,available in www.besjournal.com).
The sequences of the core and pan genes of each serogroup were then compared with the previously downloaded reference sequences (in BLAST),and filtered based on the following criteria:identity > 80%,and coverage > 80%.Gene clusters were classified according to the LPS sequence length (small to large),and the identical LPS genes of different serogroups were connected by coarse lines.In addition,functional annotation of the LPS genes was performed for each serogroup.The LPS synthesis gene structure of non-L.pneumophilawas found to be similar among non-L.pneumophilaserogroups.However,among the LPS core genes,the following differences were observed:Legionella_dumoffii_NY_23,Legionella_cherrii_DSM_19213,Legionella_shakespearei_DSM_23087,Legionella_moravica_DSM19234,andLegionella_dumoffii_Tex_KL contained more LPS genes than other non-L.pneumophilaserogroups,including lpxB_lpg2945_YP_096938.1 and lpxD_lpg2944_YP_096937.1.Furthermore,Legionella_dumoffii_NY_23,Legionella_dumoffii_Tex_KL,andLegionella_anisa_linanisettee contained the gene lpg0749_YP_094785.1,whereas other non-L.pneumophilaserogroups did not(Supplementary Figure S3,available in www.besjournal.com).
Among the 15 serogroups ofL.pneumophila,LP 14 contained the largest number of LPS genes (21 core genes and 21 pan genes),andL.pneumophilaserogroup 11 contained the fewest LPS genes (9 core genes and 11 pan genes).The genes held in common betweenL.pneumophilaand non-L.pneumophilawere found to be ostA_lpg0297_YP_094351.1,lpg0296_YP_094350.1,lpg0295_YP_094349.1,lpg2695_YP_096700.1,lpg2630_YP_096635.1,lpg2629_YP_096634.1,kdsA_lpg1182_YP_095215.1,pyrG_lpg1181_YP_095214.1,lpg0839_YP_094874.1,lpg0838_YP_094873.1,lpg0836_YP_094871.1,and CAB65217.1.
Among the non-L.pneumophilaserogroups,14-29 core LPS genes were identified,and these also had a higher distributional difference.L. pneumophilaserogroups were found to contain a higher number of LPS synthesis genes compared with the non-L.pneumophilaserogroups,and greater LPS gene differences were also detected among non-L.pneumophilaserogroups,especiallyLegionella_dumoffii_NY_23,Legionella_dumoffii_Tex_KL,andLegionella_anisa_linanisettee (Supplementary Figures S2 and S3).
The LPS genes for allLegionellastrains were downloaded from the NCBI database,and homologous alignment was subsequently performed for all samples (including 76 strains ofL.pneumophila,21 strains of non-L.pneumophila,and one strain ofCoxiella burnetii).Screening samples with the LPS reference sequence annotation,the same information with the LPS reference sequence annotation was chosen.The above results were merged into redundancy,core-pan analysis was performed based on core gene similarity,and a phylogenetic tree was constructed using the neighbor-joining method.Based on the presence of pan genes,a phylogenetic tree was constructed using the unweighted pair-group method with arithmetic means. The phylogenetic tree was constructed according to the LP grouping system using the neighbor-joining method and unweighted pair-group method. After adding the external referenceRickettsia,the results obtained were similar to those without the external reference.The analyzed data revealed a state of gradual evolution of these strains fromnon-L.pneumophilatoL.pneumophila.WZ1519012 (LP11),a strain ofL.pneumophila,represented the closest association with non-L.pneumophila,although the distribution of different serogroups ofLegionellaspecies was dispersed. According to the phylogenetic tree constructed based on core gene similarity,the evolutionary edge strains includedLegionella_shakespearei_DSM_23087_uid199258 and WD-4-1102.WD-4-1102 belongs to LP14 (Figure 1).In the phylogenetic tree based on pan gene similarity,the edge strains included SH003 andLegionella_moravica_DSM_19234_uid185635,althoughLegionella_shakespearei_DSM_23087_uid199258 and WD-4-1102 were not far apart (Figure 2).
The difference identified between the two methods was thatL.pneumophila_Paris_uid13127(LP1) andL.pneumophila_Lens_uid13126 (LP1) were clustered together in the pan tree.Whole LPS genes ofL.pneumophila_Paris_uid13127 (LP1) andL.pneumophila_Lens_uid13126 (LP1) were similar to those in other samples,although larger numbers of specific genes (264 and 223,respectively) were observed.Evolutionary trees with or without an external reference produced similar results.In the pan tree,SH003 represented the closest association with the evolutionary edge,whereas this was not the case in the core phylogenetic tree (Figure 1 and Figure 2).
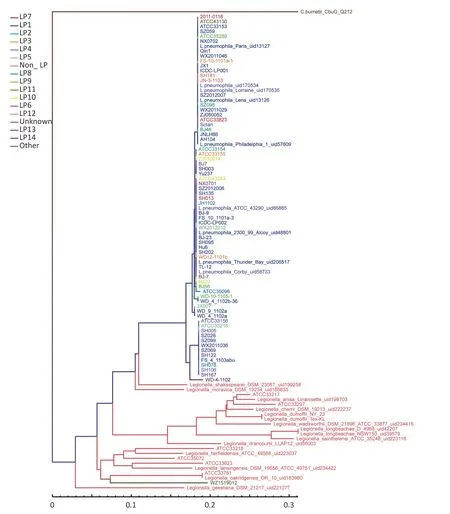
Figure 1. Phylogenetic tree based on core genes.Branch lengths are calculated from the means of the posterior probability density.Values below the nodes represent posterior probabilities.The scale bar represents substitutions per site.
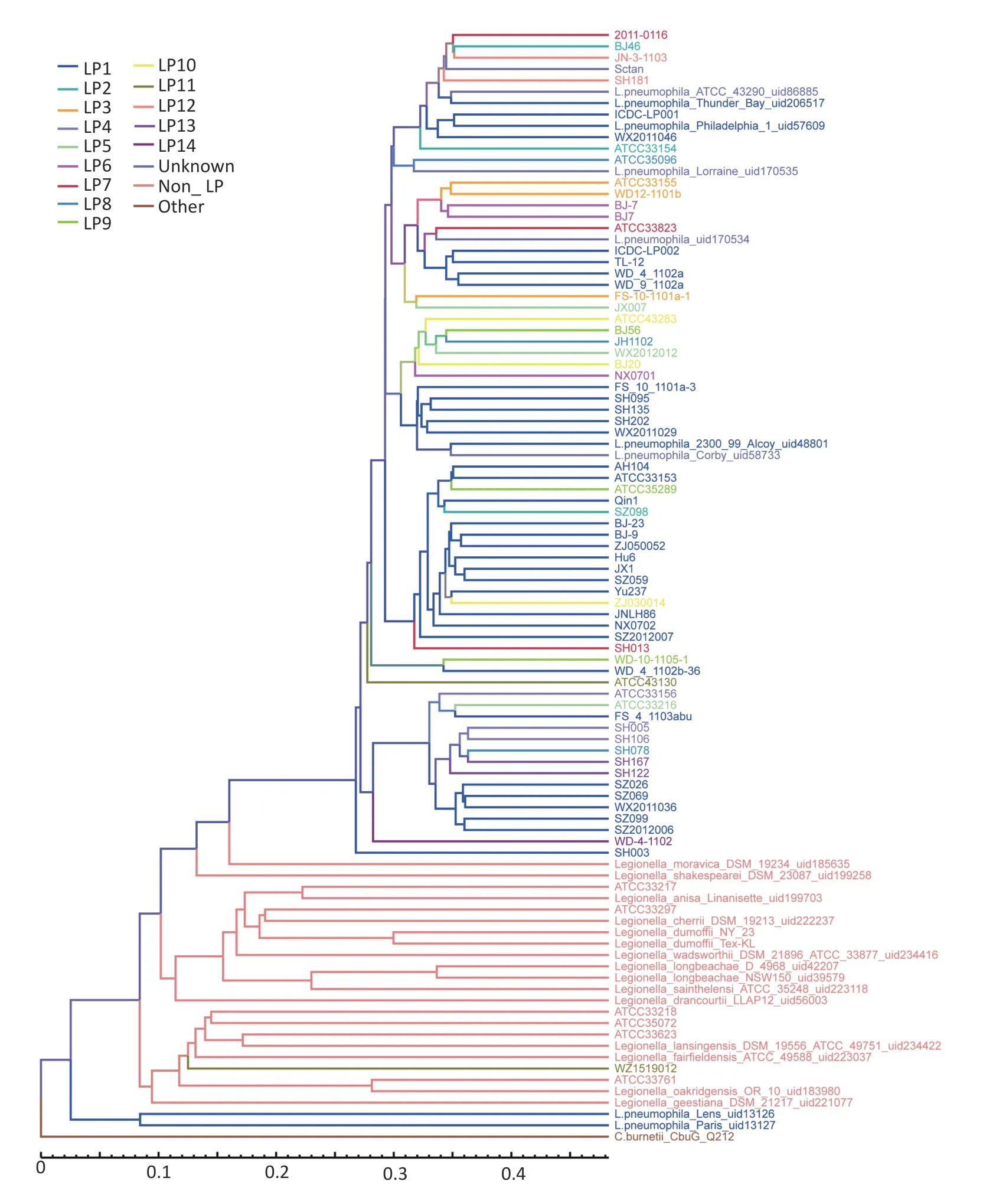
Figure 2. Phylogenetic tree based on pan genes.Branch lengths are calculated from the means of the posterior probability density.Values below the nodes represent posterior probabilities.The scale bar represents substitutions per site.
The results of phylogenetic analysis of core genes showed thatLegionella_dumoffii_NY_23,Legionella_dumoffii_Tex_KL,Legionella_cherrii_DSM_19213,andLegionella_anisa_linanisettee shared a close evolutionary relationship.Based on the phylogenetic analysis results,it was possible to speculate on how evolution may have progressed from non-L.pneumophilatoL.pneumophila.WZ1519012 (LP11)represented the closest association with non-L.pneumophila; however,the distribution of serogroups of differentLegionellaspecies was more dispersed.
Primer sequences for LP11 and LP14 were tested and screened by PCR based on O-antigen-specific genes.PCR results for hypothetical genes were eliminated in initial screening,andLegionellaserogroups 11 and 14 were chosen as the PCR templates. The O-antigen-specific genes forLegionellawere repeatedly screened by PCR.Primers were designed according to the specificwztgene in the gene cluster ofLegionellaserogroups 11 and 14.Using these primers,bands of the correct (expected)size were obtained after PCR.The PCR reaction mixture comprised,in a total volume of 30 μL the following:15 μL of PremixTaq(TaKaRa Taq Version 2 plus dye;Takara Biotechnology Co.,Ltd.),1 μL of forward primer (10 μmol/L),1 μL of reverse primer(10 μmol/L),11 μL of distilled water (Invitrogen?UltraPure Distilled Water;Thermo Fisher Scientific,Inc.),and 2 μL of template (50 ng).The PCR thermocycling conditions were:95 °C for 10 min,followed by 35 cycles of 95 °C for 30 s,annealing for 45 s (for the indicated temperatures,see Supplementary Table S3 available in www.besjournal.com),and 72 °C for 1 min,followed by 72 °C for 10 min.The primer sequences for LP11 and LP14,the size of the PCR products,and the annealing temperatures are shown in Supplementary Table S3.With the exception of the positive control group,none of the other groups presented with bands of the correct size.Hence,this demonstrated that thewztgene was highly specific inLegionellaserogroups 11 and 14.
The PCR method was tested for its specificity and sensitivity using agarose gel electrophoresis.The electrophoresis conditions were as follows:the gels were run for 40 min at 100 V in 0.5× TBE (Beijing Solarbio Science &Technology Co.,Ltd).The results obtained revealed that LP11 (Figure 3A and 3C) and LP14 (Figure 3B and 3D) displayed bands for the target gene,whereas other serogroups did not.Therefore,the specificity and sensitivity of the primers of LP11 and LP14 were confirmed by agarose gel electrophoresis.
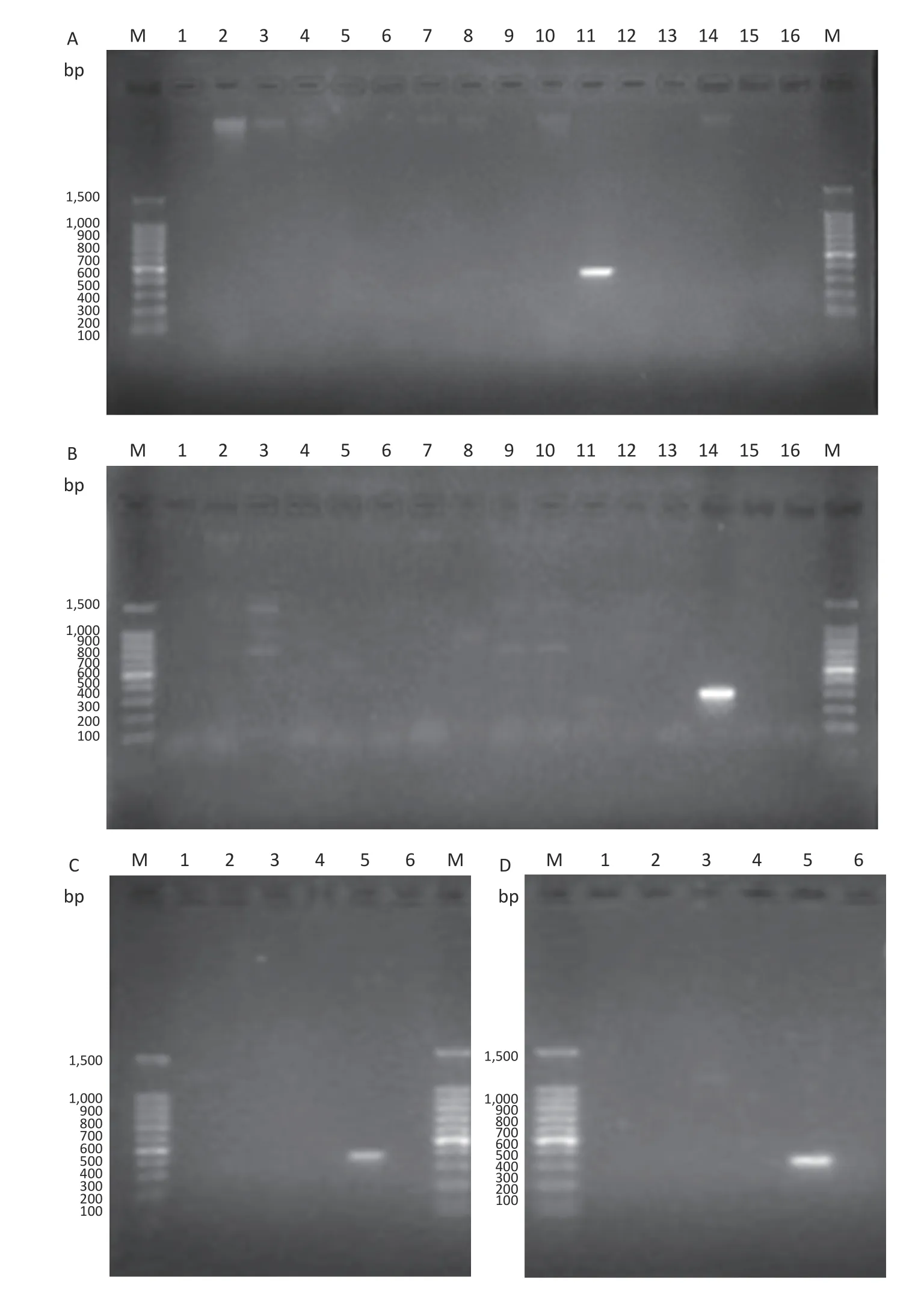
Figure 3. Agarose gel electrophoresis of PCR products.(A) LP11 PCR.PCR using primers designed for LP11:lane M,100-bp DNA marker;lanes 1-15,L.pneumophila LP1 to LP15;lane 16,negative control.(B) LP14 PCR.PCR using primers designed for LP14:lane M,100-bp DNA marker;lanes 1-15,L.pneumophila LP1 to LP15;lane 16,negative control.(C) LP11 PCR.PCR using primers designed for LP11:lane M,100-bp DNA marker;lane 1,Legionella gormanii_ATCC 33297;lane 2,Legionella dumoffii Tex-KL;lane 3,Legionella_cherrii_DSM_19213_uid 222237;lane 4,Legionella longbeachae_ATCC33462;lane 5,positive control;lane 6,negative control.(D) LP14 PCR.PCR using primers designed for LP11:lane M,100-bp DNA marker;lane 1,Legionella gormanii_ATCC 33297;lane 2,Legionella dumoffii Tex-KL;lane 3,Legionella_cherrii_DSM_19213_uid 222237;lane 4,Legionella longbeachae_ATCC33462;lane 5,positive control;lane 6,negative control.
In conclusion,the present study has shown that,based on thewztgene in the LPS cluster,it was possible to establish a rapid and specific method suitable for the identification ofL.pneumophilaserogroups.Application of this technique enabled identification of the genetic determinants of non-L.pneumophilavirulence,and allowed for important comparative studies with otherLegionellaspecies to be made.Hence,gene chip-or PCR-based methods can be applied to detect different serogroups ofLegionellaeasily and rapidly.
 Biomedical and Environmental Sciences2021年6期
Biomedical and Environmental Sciences2021年6期
- Biomedical and Environmental Sciences的其它文章
- Collaborative Efforts of Families,Schools,Health Care Providers,and the Government to Control Childhood Obesity in China
- The Improved Lipid Accumulation Product is an Accurate Index for Predicting Metabolic Syndrome in the Xinjiang Population
- Benzodiazepines and Amphetamines Use among Methadone Maintenance Participants and Their Associations with Treatment Adherence
- Quantitative Microbial Risk Assessment of Cryptosporidium and Giardia in Public Drinking Water in China*
- Campylobacter Outbreak Associated with Duck Blood Curd in 2019 in Shunyi District,Beijing,China
- Development of a High-Resolution Melting Curve Analysis to Differentiate Candida parapsilosis Complex Species
