Peptide-RNA complexation-induced fluorescence“turn on”displacement assay for the recognition of small ligands targeting HIV-1 RNA
Ling Qi ,Jiyun Zhng ,Ying Go ,Pin Gong ,Chengyun Ling ,Yo Su ,Qio Zeng ,Yfeng Zhng
a Department of Pharmacy,School of Food and Biological Engineering,Shaanxi University of Science & Technology,Xi'an,710021,China
b Xi'an Institute for Food and Drug Control,Xi'an,710054,China
Keywords:RRE RNA Rev protein Fluorescence enhancement Ligand-RNA interaction Drug screening
ABSTRACT The regulator of expression of virion(Rev)protein binds specifically to the Rev-responsive element(RRE)RNA in order to regulate the expression of the human immunodeficiency virus(HIV)-1 genes.Fluorescence indicator displacement assays have been used to identify ligands that can inhibit the Rev-RRE interaction;however,the small fluorescence indicators cannot fully replace the Rev peptide or protein.As a result,a single rhodamine B labeled Rev(RB-Rev)model peptide was utilized in this study to develop a direct and efficient Rev-RRE inhibitor screening model.Due to photon-induced electron transfer quenching of the tryptophan residue on the RB fluorophore,the fluorescence of RB in Rev was weakened and could be dramatically reactivated by interaction with RRE RNA in ammonium acetate buffer(approximately six times).The interaction could reduce the electron transfer between tryptophan and RB,and RRE could also increase RB fluorescence.The inhibitor screening model was evaluated using three known positive Rev-RRE inhibitors,namely,proflavin,6-chloro-9-[3-(2-chloroethylamino)propylamino]-2-methoxyacridine(ICR 191),and neomycin,as well as a negative drug,arginine.With the addition of the positive drugs,the fluorescence of the Rev-RRE decreased,indicating the displacement of RB-Rev.This was confirmed using atomic force microscopy(AFM)and the fluorescence was essentially unaffected by the addition of arginine.The results demonstrated that RB-Rev can be used as a fluorescent probe for recognizing small ligands that target RRE RNA.The Rev-RRE inhibitor screening model offers a novel approach to evaluating and identifying long-acting Rev inhibitors.
1.Introduction
Human immunodeficiency virus(HIV),primarily type I(HIV-1),causes acquired immune deficiency syndrome(AIDS),infecting and killing millions of people worldwide each year[1].HIV-1 is a retrovirus,and productive infection of host cells requires the reverse transcription of uncoated HIV-1 RNA into a DNA provirus,which is then integrated into the genome of the host cell[2].Current HIV-1 clinical drugs are primary inhibitors of protease and reverse transcriptase[3],and these two classes of drugs are susceptible to variability,drug resistance,and toxicity.Thus,it is urgent to identify additional new objectives[4].
The discovery of disease-causing RNA mutations has yielded a vast array of new therapeutic targets,and the growing understanding of RNA biology and chemistry has provided new RNAbased tools for the development of therapeutics[5].According to research,RNA may also be a potent drug target for the development of antiviral drugs.A number of molecular recognition steps regulate the expression of HIV genes.Specific protein/nucleic acid recognition between viral proteins and RNA sequences is an important type of interaction occurring at the gene level[6].The regulator of expression of virion(Rev)protein is one of several HIV-1-encoded regulatory proteins.mRNA entrapment in the nucleus inhibits the expression of structural genes in the absence of Rev[7].Therefore,Rev expression is required for the nuclear export of HIV structural mRNA into the cytoplasm[8].The Rev protein is a phosphoprotein with 116 amino acids and an arginine-rich domain[9],which binds to the 5'region of the HIV genome,the Rev-responsive element(RRE),identified in stem-loop IIB,one subdomain of the full-length RRE RNA[10,11].The interaction between Rev and RRE regulates mRNA transport and splicing to mediate its function[12],suggesting that the formation of Rev-RRE complexes plays a crucial role in the regulation of HIV gene expression.
The complexation of Rev and RRE RNA is a lengthy process.Small ligands that bind to the Rev binding site in RRE RNA have the potential to prevent the complexation of Rev and RRE,thereby inhibiting Rev protein function and preventing HIV-1 replication[13].Therefore,this is a new target for the treatment of HIV infection,and it is also of great importance for the screening and development of inhibitors that disrupt the interaction between protein and RNA.
Some small ligands have been reported to inhibit the complexation of Rev and RRE[14-17].Neomycin,a type of classical aminoglycoside antibiotic,can interact with RRE RNA in its lower stem region to form a specific binary complex,inhibiting the Rev-RRE complexation via an allosteric mechanism[18].Previous research demonstrated that proflavin binds to a single site on RRE IIB with a stoichiometry of 2:1 and that occupancy of this site inhibits the formation of the Rev-RRE IIB complex[19].We discovered a novel ligand,6-chloro-9-[3-(2-chloroethylamino)propylamino]-2-methoxyacridine(ICR 191),that can interact with RRE RNA at the Rev binding site and competes with the Rev peptide[20].Arginine can also bind to RRE RNA,but its binding constant is small and its affinity for binding is very weak[21].
Displacement assay is a technique for investigating the interactions between two molecules,such as ligands and receptors,by observing the signal generated by an indicator.When the indicator and test molecules compete for the same receptor binding site,the test molecules can displace the bound indicator[22].Nakatani and co-workers[23]were the first to describe a fluorescence indicator displacement(FID)assay for evaluating ligand-RNA interactions based on a novel fluorescence indicator,a 2,7-disubstituted 9Hxanthen-9-one derivative(X2S).They also synthesized a series of xanthone and thioxanthone derivatives with amino-alkoxy substituents to improve the binding selectivity of X2S[24].Similarly,we used ICR 191 as a fluorescence indicator in a ratiometric FID assay to evaluate the interaction of antagonists with RRE RNA at the Rev binding site[20].We also developed a graphene-oxide-enhanced and proflavine-probed assay for evaluating ligand-RNA interactions[25].In the FID assay for ligand-RNA interactions,the fluorescence of the indicator is typically quenched(“turned off”)when the ligand binds to the receptor.Upon displacement of the bound indicator with test molecules,the fluorescence emitted by the free unbound indicator is recovered(“turned on”).
Identifying a small-molecule compound that emits fluorescence and binds to RRE RNA at the Rev binding site to serve as a fluorescence indicator in the FID assay is relatively challenging.Due to the substantial differences between their chemical structures and binding affinities,small-molecule fluorescence indicators cannot completely replace the function of peptides or proteins.Fluorescently-labeled peptides have been utilized to evaluate and screen small inhibitors that bind to HIV-1 RNAs,for instance,in fluorescence polarization/anisotropy[26-28]or fluorescence resonance energy transfer(FRET)studies[29,30].However,RNA can affect the fluorescence of fluorescently-labeled peptides,leading to nonuniformity in the fluorescence polarization assay;matching the donor-receptor pair in the FRET study could be challenging.Utilizing the self-fluorescence of fluorescently-labeled peptides would be much simpler.
A Rev peptide containing the arginine-rich region has been shown to bind specifically to the stem-loop IIB of RRE RNA[31,32],and we discovered that the single amino acid tryptophan(Trp)in the Rev peptide may play a unique role because it is known to serve as an electron donor and efficiently quench oxazine and rhodamine fluorophores via photon-induced electron transfer(PET)[33].It was demonstrated that these fluorophores coupled to the N-terminus of a peptide are effectively quenched by the Trp in the same peptide[34].Consequently,a Rev peptide labeled with a commercial rhodamine B(RB)dye(RB-Rev;Fig.1A)was employed and its viability as a direct fluorescent probe was evaluated.The fluorescence of RB-Rev was weak,but it could be augmented by interacting with the 34-nucleotide model RRE RNA(Fig.1B).The fluorescence was then diminished by adding a known positive Rev inhibitor(Fig.1C).Using RB-Rev as the fluorescent probe,we aimed to develop a direct and efficient fluorescence“turn on”Rev-RRE inhibitor screening model.Fig.1D depicts the chemical structures of the small-molecule inhibitor models used in this study.
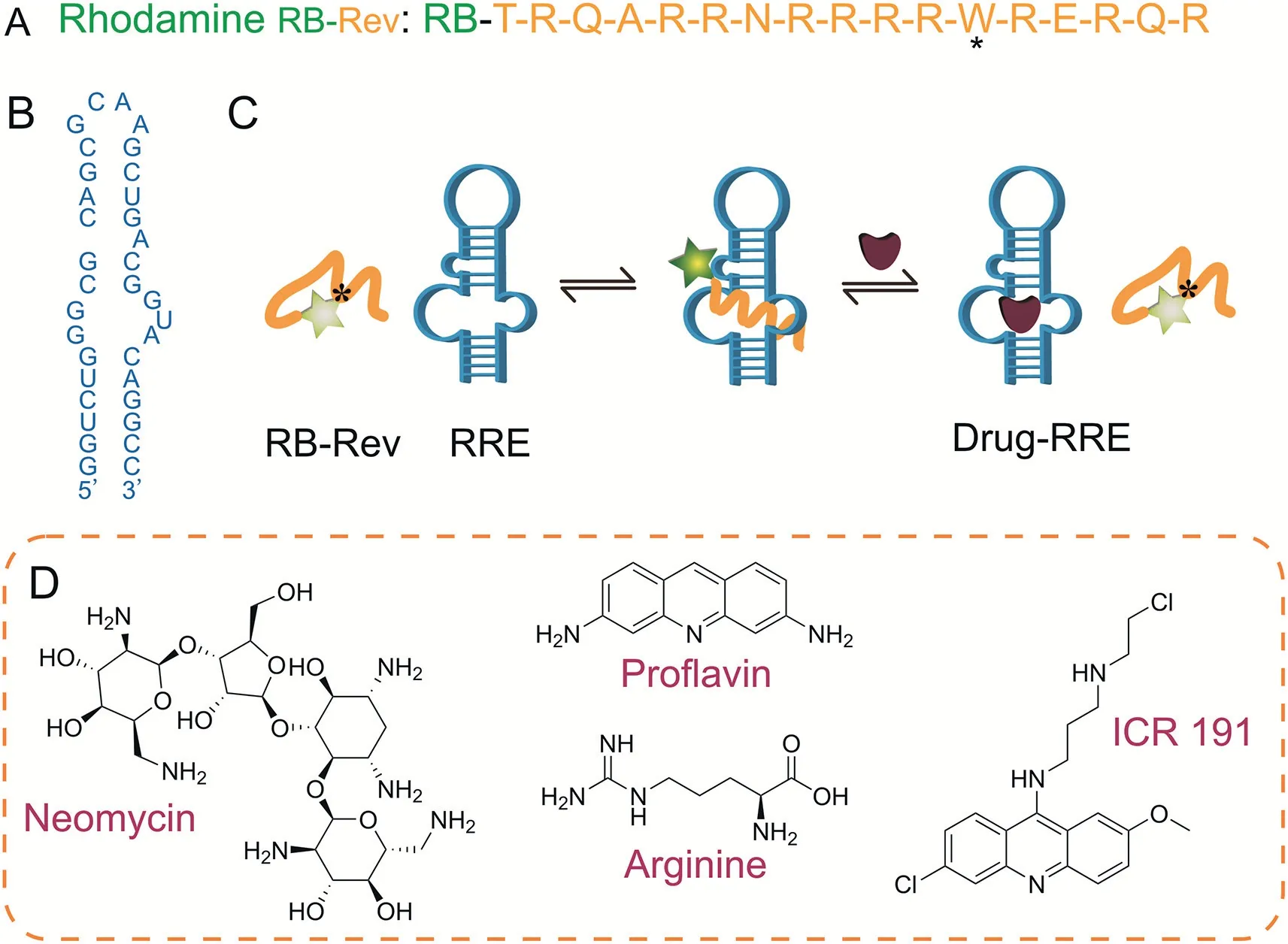
Fig.1.The sequences and structures of the molecules used in this study and the scheme of Rev-RRE inhibitor screening.(A)The sequence of rhodamine B labeled regulator of expression of virion(RB-Rev)peptide model amino acid(W:tryptophan).(B)The sequence of the Rev-responsive element(RRE)RNA model nucleotide.(C)Illustration of the peptide-RNA complexation-induced fluorescence“turn on”displacement assay for the recognition of small ligands targeting RRE RNA.(D)The chemical structures of neomycin,proflavin,arginine,and 6-chloro-9-[3-(2-chloroethylamino)propylamino]-2-methoxyacridine(ICR 191)used in this study.
2.Materials and method
2.1.Materials
RB-Rev,variant RB-Rev,and HIV-1 RRE RNA were customsynthesized and HPLC-purified by Sangon Biotech Co.,Ltd.(Shanghai,China).ICR 191 dihydrochloride was purchased from Sigma-Aldrich Co.,Ltd.(St.Louis,MO,USA),while neomycin sulfate was purchased from Meilun Biotechnology Co.,Ltd.(Dalian,China).Proflavin and arginine were purchased from Macklin Biotechnology Co.,Ltd.(Shanghai,China).Additionally,RB was purchased from Aladdin Biotechnology Co.,Ltd.(Shanghai,China).Tryptophan was obtained from Solarbio Science & Technology Co.,Ltd.(Beijing,China).The remaining reagents were all of the analytical grade,commercially available,and employed without further purification.
RNase-free tubes and tips were purchased from Thermo Science& Technologies Co.,Ltd.(Waltham,MA,USA),and diethylpyrocarbonate(DEPC)water was obtained from Phygene Biotechnology Co.,Ltd.(Fuzhou,China).The RNA solutions were prepared using DEPC water to prevent nuclease contamination.
2.2.Fluorescence measurements
In the binding assay,the fluorescence spectra were measured by holding constant the concentrations of RB-Rev and RB while varying the concentrations of RNAs,as well as by holding constant the mole ratio of RB-Rev and RRE RNA while varying the concentrations of RB-Rev and RRE RNA.The peptide-RNA complex was thoroughly mixed with ammonium acetate buffer solution and incubated at 37°C for 10 min.
During drug displacement experiments,the fluorescence spectra were observed by maintaining a constant concentration of the RB-Rev-RRE complex while adding model drugs in varying concentrations.The solutions were incubated for 10 min at 37°C in a buffer containing ammonium acetate,and the fluorescence spectra were measured.
All fluorescence spectra were measured using a Cary Eclipse Fluorescence Spectrophotometer(Agilent Technologies Co.,Ltd.,Santa Clara,CA,USA)and a quartz cuvette with a path length of 0.2 cm.The excitation wavelengths of RB-Rev and RB were set to 555 nm.The emission spectra of RB-Rev and RB were recorded from 565 to 650 nm.Emission and excitation slits were both set to 5 nm,the scanning speed was set to 600 nm/min,and the photomultiplier voltage was set to a high value.The final volume of the reaction solution was 200μL.All fluorescence spectra were recorded at least three times.
The relative fluorescence quantum yields of RB-Rev(1μM)and RB-Rev-RRE(1μM)were measured using RB(1μM)as a reference standard.The excitation wavelength was set to 555 nm,and the emission spectrum scanning range was 565-650 nm.The UV-vis absorbance measurements were performed on a P4 UV-vis spectrophotometer(Mapada Instrument Co.,Ltd.,Shanghai,China)using a quartz cuvette(500μL).
The formula for calculating the fluorescence quantum yields of RB-Rev and RB-Rev-RRE is as follows:
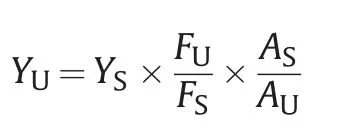
where,YUis the quantum yield of RB-Rev or RB-Rev-RRE,YSis the fluorescence quantum yield of RB,FUis the integrated fluorescence intensity of RB-Rev or RB-Rev-RRE,FSis the integrated fluorescence intensity of RB,AUis the absorbance value of RB-Rev or RBRev-RRE,andASis the absorbance value of RB.
2.3.Atomic force microscopy(AFM)
The RB-Rev(0.5μM)-RRE RNA(0.5μM)complex was preformed,followed by the addition of three Rev-RRE model inhibitors(ICR 191,neomycin,and proflavin).All samples were incubated for 10 min at 37°C in an ammonium acetate buffer solution(pH 7.0,10 mM),after which 1μL of the reaction solution was deposited on an ethanol-cleaned coverslip and dried under a gentle flow of nitrogen.Ultimately,the coverslip containing the sample was mounted on the AFM scanner(NSK Ltd.,Tokyo,Japan)for measurement.
3.Results and discussion
3.1.Optimization of fluorescence enhancement conditions
The experimental conditions for the binding of RB-Rev to RRE RNA,including the buffer solution type,RB-Rev concentration,and mole ratio,were optimized for maximum fluorescence enhancement.
3.1.1.Types of buffer solution
In four different buffer solutions,the fluorescence of RB-Rev and RB-Rev-RRE complex(both at 0.5μM)was measured:ammonium acetate buffer(pH 7.0,10 mM),Tris-HCl buffer(pH 6.8,10 mM),2-(N-morpholino)ethanesulfonic acid(MES)buffer(pH 7.0,10 mM),and phosphate buffer(pH 6.8,10 mM).At the emission wavelength(585 nm)of RB-Rev,the results demonstrated that the greatest fluorescence enhancement was obtained in ammonium acetate buffer(approximately 6 times)(Fig.2A),and at the same wavelength,the fluorescence enhancement factors in MES,Tris-HCl,and phosphate buffer were approximately 3.9,3.2,and 3.8,respectively(Figs.2B-D).Therefore,ammonium acetate buffer was chosen for subsequent experiments.
3.1.2.Concentration of RB-Rev and RRE RNA
RRE RNA was added to different concentrations of RB-Rev(0.1,0.3,0.5,0.7,and 1.5μM)at a mole ratio of 1:1 in ammonium acetate buffer to determine the optimal concentration of RB-Rev(Fig.3).As the concentrations of reactants increased,more fluorescent complexes were produced.However,the self-fluorescence intensity of RB-Rev did not significantly increase as its concentration increased.Thus,the fluorescence enhancement factors continued to rise;a remarkable fluorescence enhancement factor of approximately 6 was already observed when the concentration of reactants was 0.5μM.

Fig.2.Fluorescence spectra of rhodamine B labeled regulator of expression of virion(RB-Rev)(0.5μM)in the absence and presence of Rev-responsive element(RRE)RNA(0.5μM)in(A)ammonium acetate buffer,(B)2-(N-morpholino)ethanesulfonic acid(MES)buffer,(C)Tris-HCl buffer,and(D)phosphate buffer.
3.1.3.Mole ratio of RB-Rev and RRE RNA
The optimal mole ratios of RB-Rev and RRE RNA were also investigated.RB-Rev(0.5μM)was mixed with different concentrations of RRE RNA(0,0.25,0.5,0.75,and 1μM)at different mole ratios(1:0,1:0.5,1:1,1:1.5,and 1:2),and fluorescence was measured(Fig.4).As the concentration of RRE RNA increased,so did the fluorescence intensity.However,the fluorescence enhancement factor did not change significantly once the RRE RNA concentration exceeded 0.5μM.
3.2.The Rev-RRE inhibitor screening model
The combination of RB-Rev and RRE RNA at a theoretical mole ratio of 1:1 resulted in nearly all RRE molecules existing as the RBRev-RRE complex.In addition,equal amounts of RB-Rev and RRE can guarantee that the test drugs interact directly with the complex to displace RB-Rev during the screening process.This can result in direct fluorescence changes,and a lower complex concentration typically necessitates a lower test drug dose.Therefore,a concentration ratio of 1:1 between RB-Rev and RRE RNA was chosen,with both concentrations being 0.5μM.If a test drug could reduce the fluorescence of the RB-Rev-RRE complex,it would be considered as a positive inhibitor.
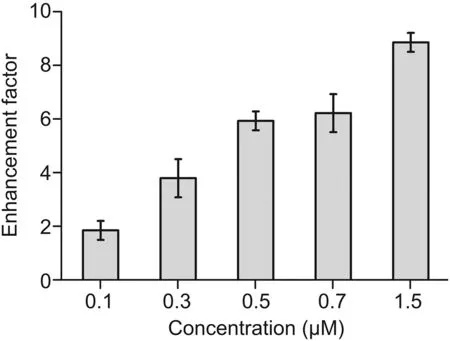
Fig.3.Fluorescence enhancement factor histogram of different concentrations of rhodamine B labeled regulator of expression of virion(RB-Rev)(0.1,0.3,0.5,0.7,and 1.5μM)bound to Rev-responsive element(RRE)RNA at a mole ratio of 1:1 in ammonium acetate buffer.
3.3.Evaluation of Rev-RRE inhibitor screening
By adding RB-Rev as a fluorescent probe and observing the fluorescence variations of the RB-Rev-RRE complex,three known positive Rev inhibitors,proflavin,ICR 191,and neomycin,were used to verify the viability of a drug screening model for identifying Rev-RRE inhibitors.Arginine was chosen as a negative model drug because the Rev protein contains an arginine-rich binding domain,but a single arginine binds to RRE RNA with a very weak affinity.The RB-Rev(0.5μM)and RRE RNA(0.5μM)complex was first prefabricated and then mixed with varying concentrations of proflavin(0,0.5,1,3,5,10,15,20,and 40μM),ICR 191(0,0.5,1,3,5,7.5,and 10μM),neomycin(0,0.5,1,2,3,4,and 5μM),and arginine(0,2,4,and 5μM).As the concentrations of the three positive drugs increased,the fluorescence intensity of the complex decreased(Figs.5A-C),indicating that the three drugs could replace RB-Rev.The concentration(IC50)of proflavin,ICR 191,and neomycin was approximately 14.0,6.3,and 2.3μM,respectively,when the displacement rate was 50%.The results indicated that neomycin possessed the strongest displacement ability,followed by ICR 191 and proflavin.Due to its low binding affinity,arginine was unable to replace RB-Rev,and the fluorescence intensity remained nearly unchanged(Fig.5D).As a control,the fluorescence spectra of four model drugs were also measured,and the model drugs exhibited no background fluorescence at the excitation wavelength of RB-Rev(Fig.S1).The results could suggest that the release of RB-Rev caused by the addition of the three positive drugs was the primary cause of the decrease in fluorescence observed in the RB-Rev displacement assay.
It has been reported that AFM can measure the adhesion force between Rev and RRE[35],so we utilized AFM to confirm the competition between positive drugs and RB-Rev.The AFM images revealed that the morphology of the Rev-RRE complex(0.5μM)was broad and tall(Fig.S2A),but the length and height were both reduced after the complex was incubated with ICR 191(10μM),neomycin(3μM),or proflavin(40μM)(Figs.S2B-D),indicating dissociation of the Rev-RRE complex.This was consistent with the experimental results of the fluorescence assay.Thus,using RB-Rev as a fluorescent probe,the drug screening model can be used to identify positive inhibitors that disrupt the interaction between Rev and RRE RNA.

Fig.4.Fluorescence spectra of rhodamine B labeled regulator of expression of virion(RB-Rev)(0.5μM)after incubation with increasing concentrations of Rev-responsive element(RRE)RNA(0,0.25,0.5,0.75,and 1μM).The inset shows the concentrationdependent curve of RB-Rev incubating with RRE RNA.
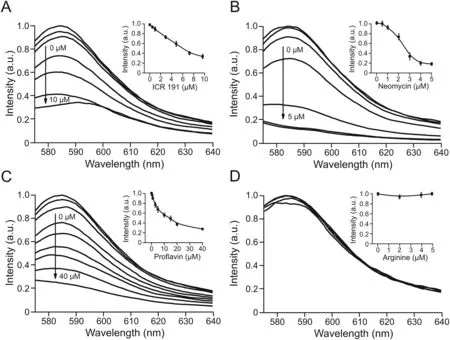
Fig.5.Fluorescence spectra of the preformed rhodamine B labeled regulator of expression of virion(RB-Rev)(0.5μM)-Rev-responsive element(RRE)RNA(0.5μM)complex after incubation with increasing concentrations of(A)6-chloro-9-[3-(2-chloroethylamino)propylamino]-2-methoxyacridine(ICR 191)(0,0.5,1,3,5,7.5,and 10μM),(B)neomycin(0,0.5,1,2,3,4,and 5μM),(C)proflavin(0,0.5,1,3,5,10,15,20,and 40μM),and(D)arginine(0,2,4,and 5μM).The inset shows the concentrationdependent curve of the complex incubating with ICR 191,neomycin,proflavin,and arginine,respectively.
3.4.The mechanism involved in fluorescence variation
The relative quantum yields of RB-Rev and RB-Rev-RRE were determined.Using RB as a standard,with a quantum yield in ethanol of 71%[36],the quantum yields of RB-Rev and RB-Rev-RRE in ammonium acetate buffer were calculated to be 1.53% and 14.48%,respectively.After combining with RRE RNA,the fluorescence quantum yield of RB-Rev increased by approximately nine folds.This was consistent with previous experimental results of the interaction between RB-Rev and RRE.
The effect of Trp on the fluorescence of a single RB molecule was investigated,and it was observed that the fluorescence of RB(100 nM)decreased over time after mixing with Trp(10μM)(Fig.S3).The experimental outcome could further validate the PET interaction between free Trp and rhodamine fluorophore[33].
The effect of pH on the fluorescence of RB-Rev was then investigated.It was discovered that the fluorescence of RB-Rev increased under acidic(pH 3)conditions(Fig.6).When the solution pH was below the isoelectric point of Trp(5.89),the electrostatic repulsion between the cationic RBH+(pKa 3.1)and TrpH+was likely to reduce the intermolecular electron transfer interaction,resulting in stronger fluorescence.In another assay,the fluorescence of RB-Rev and a variant RB-Rev in which the Trp residue was removed was compared.At the same concentration,the fluorescence of variant RB-Rev was stronger than that of RB-Rev(Fig.S4),indicating that Trp in the Rev peptide inhibited RB fluorescence.
The fluorescence intensity of RB(0.1μM)increased as the concentration of RRE RNA increased(0,0.25,and 1μM)(Fig.S5).Therefore,it is believed that RRE may also increase the fluorescence of RB.
For this screening model,the fluorescence variation mechanism can be determined.Through PET,the Trp residue of the Rev peptide induced the quenching of the RB fluorophore.Upon binding to the RRE,the Rev adopted a new conformation,the Trp-RB electron transfer interaction was reduced,and the RRE could also boost the fluorescence of RB,resulting in the final fluorescence enhancement.The addition of positive inhibitors dislodged the Rev peptide from the RRE,resulting in a decrease in fluorescence.

Fig.6.Fluorescence spectra of rhodamine B labeled regulator of expression of virion(RB-Rev)(1μM)in aqueous solutions with different pH values.
4.Conclusions
Using RB-Rev as a fluorescent probe,a direct and efficient Rev-RRE inhibitor screening model was created.The interaction between RB-Rev and RRE RNA resulted in different fluorescence enhancement effects in different buffer solutions,and the fluorescence enhancement factor was significant when their concentration ratio was theoretically 1:1.Interaction experiments between known positive and negative model drugs and the prefabricated RB-Rev-RRE complex demonstrated the feasibility of the drug screening model,which was also confirmed by AFM.The Trp residue of Rev could quench the RB fluorophore via PET,the interaction with RRE could reduce the electron transfer between Trp and RB,and RRE could also boost the fluorescence of RB.This screening model may be used to study the interactions of RRE RNA with other Rev inhibitors and may provide a novel method for studying ligand-RNA interactions.
CRediT author statement
Liang Qi:Conceptualization,Methodology,Writing-Reviewing and Editing,Visualization,Project administration,Funding acquisition;Jiayun Zhang:Investigation,Formal analysis,Writing-Original draft preparation;Ying Gao:Investigation;Pin Gong:Resources;Chengyuan Liang:Resources,Supervision;Yao Su:Investigation;Qiao Zeng:Resources,Supervision;Yafeng Zhang:Conceptualization,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of Shaanxi Province of China(Grant No.:202012119),the Start-up Funding of Shaanxi University of Science and Technology(Grant No.:2019BJ-48),and the Innovation Capability Support Program of Shaanxi Province of China(Grant No.:2021PT-044).Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.07.003.
 Journal of Pharmaceutical Analysis2022年6期
Journal of Pharmaceutical Analysis2022年6期
- Journal of Pharmaceutical Analysis的其它文章
- Potential use of a dried saliva spot(DSS)in therapeutic drug monitoring and disease diagnosis
- Rabdosia serra alleviates dextran sulfate sodium salt-induced colitis in mice through anti-inflammation,regulating Th17/Treg balance,maintaining intestinal barrier integrity,and modulating gut microbiota
- Identification of potential anti-pneumonia pharmacologicalcomponents of Glycyrrhizae Radix et Rhizoma after the treatment with Gan An He Ji oral liquid
- LC-MS/MS method for the quantitation of serum tocilizumab in rheumatoid arthritis patients using rapid tryptic digestion without IgG purification
- Discovery of pulmonary fibrosis inhibitor targeting TGF-βRI in Polygonum cuspidatum by high resolution mass spectrometry with in silico strategy
- Development of a radiolabeled site-specific single-domain antibody positron emission tomography probe for monitoring PD-L1 expression in cancer
