Epigenetic Repression of SATB1 by Polycomb Group Protein EZH2 in Epithelial Cells△
Lei Li,Lu Lu,Xiang Lü,Xue-song Wu,De-pei Liu*,and Chih-chuan Liang
National Laboratory of Medical Molecular Biology,Institute of Basic Medical Sciences,Chinese Academy of Medical Sciences &Peking Union Medical College,Beijing 100005,China
POLYCOMB group (PcG) proteins are divided into two biochemically characterized classes of nuclear regulators,forming the Polycomb repressive complexes 1 and 2 (PRC1 and PRC2).PRC2 could initiate gene silencing by catalyzing trimethylation of histone H3 on lysine 27 (H3K27) with its site-specific methyltransferase activity.1-4Trimethylated H3K27 (H3K-27me3) then serves as a docking site for further recruitment of PRC1 and other repressive factors to maintain gene silencing during certain stages of body development or cell differentiation.5,6PRC2 consists of three core components,EED,SUZ12,and EZH2,among which EZH2 is the central one with enzyme activity.Recent studies in mammalian cells have identified a large variety of PRC2 target genes known to be involved in key pathways controlling embryonic development,cell differentiation,stem cell biology,and cell fate determination.7-9
SATB1 (special AT-rich sequence binding protein 1) is a T-cell-enriched gene regulator,which functions as a chromatin organizer by periodically binding to specific matrix attachment regions and tethering them to the nuclear matrix.10,11SATB1 can also directly recruit chromatin modifying factors to orchestrate expressions of a large number of genes.12,13It has been reported that SATB1 contributes to compacting chromatin into dense loop domains and controlling the expression of cytokine genes in Th2 cells and MHC class I genes in Jurkat cells.14,15A recent study also reported the regulatory effect of SATB1 on embryonic stem cell differentiation and Nanog expression.16Apart from its roles in regulating numerous genes in T cells,a previous study demonstrated that SATB1 expressed during the early stage of erythroid differentiation could modify globin gene expression.17Furthermore,our recent findings suggested that SATB1 is a critical player contributing to transcriptionally active looping events in human β-globin gene cluster in K562 cells and such regulatory effects are exerted through matrix related nuclear relocation by its nuclear matrix targeting sequences.18,19
Given the functions of SATB1 as a global gene regulator,the regulation of its own expression is of importance to further understand its biological roles.Previous studies have shown that SATB1 expression is regulated by interleukin-4 in human differentiating Th2 cells.20,21However,the mechanisms underlying the expression pattern of SATB1 in non-T or non-erythroid cells still remain largely unknown.Different from T cells or erythroid cells,which derive from common ancestor cells during hematopoiesis,cells from other lineages express SATB1 at lower levels.15,22As SATB1 also contains conserved homeobox domain,we assumed that SATB1 might also be a PcG target gene as Hox genes.Therefore,in this study we explored the potential regulatory mechanisms underlying the repression of SATB1 gene expression with HeLa epithelial cells.
MATERIALS AND METHODS
Cell culture and drug treatment
HeLa cells [Peking Union Medical College (PUMC) Culture Collection]were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen,Carlsbad,CA,USA) supplemented with 10% fetal bovine serum (FBS).K562 cells and Jurkat cells (PUMC Culture Collection) were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS for later use as control cells in the chromatin immunoprecipitation (ChIP) assay.
PRC2 initiates silencing of target genes by modifying H3K27 with trimethylation mark,during which histone deacetylase (HDAC) activity is required for efficient repression of target genes.5,23Furthermore,the repression process will generally lead to DNA methylation within promoter regions of target genes.24,25Therefore,it is reasonable to suppose that inhibition of DNA methylation or HDAC activity could induce expression of PRC2 target genes,which is the basis of the following treatment.
HeLa cells were treated continuously for 4 days with DNA methylation inhibitor 5-Aza-C (Sigma-Aldrich,St.Louis,MO,USA) dissolved in dimethyl sulfoxide (DMSO)to a concentration of 5 μmol/L to“dilute”DNA methylation during cell divisions.HDAC inhibitor trichostatin A(TSA) (Sigma-Aldrich) was dissolved in DMSO and two different concentrations (0.5 μmol/L and 1 μmol/L) were used separately to treat HeLa cells for 16 hours.The control group was treated with the same amount of DMSO alone.
Plasmid construction
Full-length EZH2 coding sequence was amplified from HeLa cDNA with polymerase chain reaction (PCR) and ligated into pcDNA3.1 (Invitrogen).The three fragments of SATB1 promoter region were PCR-amplified from HeLa genomic DNA and ligated into pGL3-basic (Promega,Madison,WI,USA) atKpnIandXhoIsites to form pGL3-SATpro.The primers are as follows:5’-CAGTggtaccGCCAGGGCGACTCTAGAG-3’ (forward strand for 14-1222 fragment),5’-CAGTggtaccGGCAGGGAGAGGGTCCT-3’ (forward strand for 327-1222 fragment),5’-CAGTggtaccGGAGAGACGCGAAGAGACCT-3’ (forward strand for 663-1222 fragment),and 5’-AGCTctcgagCACTTCAAAACTTGACAGCACATA-3’ (reverse strand primer).Sequence accuracy was confirmed by means of DNA sequencing.
Luciferase assay
HeLa cells were plated at 0.5×104-1×104cell/well in 24-well plates.For testing the promoter activities of different SATB1 promoter segments,some plated HeLa cells were transfected with 100 ng pGL3-SATpro plasmids or the negative control pGL3-basic plasmids.The phRL-TK plasmids were added into all the wells as internal control for luciferase assay.
To test the effects of EZH2 on SATB1 promoters,some plated HeLa cells were transfected with 100 ng pGL3-SATpro plasmids or the negative control pGL3-basic plasmids along with pcDNA3.1 or EZH2 expression plasmids.The phRL-TK plasmids were added in each well as internal control.The transfection reagent applied in this procedure is Lipofectamine 2000 (Invitrogen).
Luciferase assays were performed with Dual Luciferase Reporter Assay Kit (Promega) on the cells harvested 48 hours after transfection.
Stable RNAi of EZH2
The targeting sequence (passenger strand) for EZH2 knockdown is 5’-ATATGACTGCTTCCTACAT-3’.The short hairpin RNA (shRNA) oligos were ligated into pSirenRetroQ(Clontech,Mountain View,CA,USA) atBamHIandEcoRIsites to generate pSiren-shEZH2.The construct pSirenshLuc expressing shRNA against luciferase was used as control.The two types of plasmids were separately trasnfected into GP293 cells along with pVSV-G (Clontech)to generate retroviruses.Sixty hours after transfection,the media were collected,filtered,and stored at-80°C for later use.Forty-eight hours after infecting HeLa cells with the media,1 μg/mL puromycin was added and the cells were maintained for 7-10 days to generate stable knockdown cell lines.
Quantitative reverse transcription-PCR (RT-PCR)
Total RNA of HeLa cells was isolated by using TRIzol reagent (Invitrogen) and reverse transcription was performed following the manufacturer’s manual (New England Biolabs,Ipswich,MA,USA).Primer sequences are as follows:EZH2,5’-GCCAGACTGGGAAGAAATCTG-3’ (forward),5’-TGTGtTGGAAAATCCAAGTCA-3’ (reverse);HoxA9,5’-AGCGGTGCCCCTATACAAAAC-3’ (forward),5’-TGCCTCTCGGTGAGGTTGAG-3’ (reverse) (positive control);SATB1,5’-GATCTATGAATAAGCCTTTGGAG-3’ (forward),5’-TTTCGTCCTGGTATATTCGGT-3’ (reverse);GAPDH,5’-GGTGAAGGTCGGAGTCAACGGA-3’ (forward),5’-GAGGGATCTCGCTCCTGGAAGA-3’ (reverse) (internal control).
Western blot
HeLa cells from EZH2 over-expression,EZH2 RNAi,or control groups were lysed with RIPA buffer [50 mmol/L Tris-HCl (pH 8.0),150 mmol/L NaCl,1% NP-40,0.5%sodium deoxycholate,0.1% sodium dodecyl sulfate (SDS),protease inhibitor cocktail from Sigma-Aldrich]and the extracts were sonicated for 15 seconds using a Branson 150 sonicator (Branson Ultrasonics,Danbury,CT,USA)before blotting.The blots were probed by anti-EZH2 (Cell Signaling Technology,Boston,MA,USA),anti-H3K27me3(Upstate,Billerica,MA,USA),anti-H3 (Abcam,Cambridge,MA,USA),and anti-tubulin (Abcam) antibodies.
ChIP assays
ChIP assays were performed as described in the protocol by Upstate with a few minor modifications.After crosslinking with formaldehyde,isolated nuclei of HeLa,K562,and Juarkat cells were lysed in SDS lysis buffer [1% SDS,10 mmol/L EDTA,and 50 mmol/L Tris (pH 8.1)]and sonicated with a Branson 150 sonicator down to sizes of 200-750 bp.Antibodies against EZH2 (Cell Signaling Technology) and H3K27me3 (Upstate) were used for immunoprecipitation.IgG was employed as a negative control to EZH2 and H3K27me3 antibodies.PCR primers used in this procedure are as follows:HoxA9-B (proximal promoter),5’-TCGCCAACCAAACACAACAGTC-3’ (forward),5’-AAGCAGCCAAATCGCATTGT-3’ (reverse) (positive control of PRC2 target genes);HoxA9-C (first intron),5’-GCAAAAGACGAGTGATGCCA-3’ (forward),5’-AGCCTACCATCAACAGTTGTGC-3’ (reverse) (positive control);SATB1,5’-ACGTCGTTTCCCCAGCAC-3’ (forward),5’-AAACGTCTAGAAGAGTAGCCATGAGAA-3’ (reverse);GAPDH,5’-TACTAGCGGTTTTACGGGCG-3’ (forward),5’-TCGAACAGGAGGAGCAGAGAG-3’ (reverse) (negative control).
RESULTS
Inhibition of DNA methylation or HDAC activity increases transcription of SATB1 gene in HeLa cells
HeLa epithelial cells were chosen in the present study to test whether SATB1 is a PRC2 target gene in non-T or non-erythroid cells.After 4-day continuous treatment with DNA methylation inhibitor 5-Aza-C,the transcription of SATB1 in HeLa cells increased compared to the control group as shown by the results of quantitative RT-PCR (Fig.1A).The transcription level of a well studied PRC2 target gene HoxA9 was also elevated,which serves as a positive control,while the transcription of EZH2 remained unchanged (Fig.1A).
HeLa cells were also treated with 0.5 μmol/L and 1 μmol/L of TSA to test if inhibition of HDAC activity induces SATB1 expression.The results of quantitative RT-PCR showed that TSA treatment at 1 μmol/L upregulated SATB1 as well as HoxA9 expression (Fig.1B).In addition,EZH2 transcription was slightly decreased after treatment with 1 μmol/L of TSA (Fig.1B).These findings suggest that PRC2 probably participate in repressing SATB1 gene.
EZH2 represses SATB1 gene expression in HeLa cells but not K562 or Jurkat cells
To test if PRC2 directly regulates the expression of SATB1 gene,we cloned three proximal promoter regions of SATB1(Fig.2A) and detected their activities in response to expression of exogenous EZH2.Luciferase assay showed that all the three regions exhibited promoter activities compared with negative control pGL3-basic,and the promoter activity gradually decreased as the length shortened (Fig.2B).The three fragments were then co-transfected with EZH2 expression vectors or control vectors pcDNA3.1 into HeLa cells.The results of luciferase assay showed that over-expression of EZH2 reduced activities of all the three fragments by about 50% (Fig.2C).
In HeLa cells over-expressing EZH2 as confirmed by immunoblot,SATB1 mRNA level was observed to decrease by about 70% (Fig.3A).It was noted that global H3K27me3 level did not change apparently (Fig.3A),indicating that the repressive effect of EZH2 might be restricted to specific individual genes.
Then we measured SATB1 expression level in response to knockdown of EZH2.As confirmed by Western blot and quantitative RT-PCR,the EZH2 level was decreased by about two-thirds (Fig.3B).The decrease of global H3K27me3 further confirmed RNAi of EZH2 (Fig.3B).Knockdown of EZH2 in HeLa cells resulted in a three-fold increase of SATB1 expression compared to control RNAi cells (Fig.3B).That effect was not observed in K562 cells or Jurkat cells,SATB1 level was even slightly reduced in EZH2 RNAi K562 cells (Figs.3C,D).
Taken together,EZH2 may contribute to repression of SATB1 in HeLa cells but not in K562 cells or Jurkat cells.
Repression of SATB1 gene by EZH2 is mediated by epigenetically catalyzing the formation of H3K27-me3
EZH2-containing PRC2 complex initiates gene silencing by epigenetically catalyzing the formation of H3K27me3,so we detected EZH2 recruitment and histone modification status on SATB1 proximal promoter in HeLa cells,K562 cells,and Jurkat cells.ChIP assay using anti-EZH2 antibody showed recruitment of EZH2 to SATB1 promoter in HeLa cells,with EZH2 recruitment on proximal promoter and first intron of HoxA9 served as positive control,and GAPDH promoter as negative control (Fig.4).In contrast to the results in HeLa cells,no apparent EZH2 recruitment was detected in K562 or Jurkat cells (Fig.4).ChIP assay using anti-H3K27me3 antibody showed enriched H3K27 trimethylation on SATB1 promoter as well as on HoxA9 promoter in HeLa cells,but not in K562 cells or Jurkat cells (Fig.4),consistent with the results in respect of EZH2 recruitment on the same regions.These findings confirmed that repression of SATB1 gene by EZH2 in HeLa cells is mediated by H3K27me3 modification.

Figure 1.5-Aza-C and trichostatin A (TSA) treatments increase SATB1 expression in HeLa cells as demonstrated by quantitative reverse transcription-polymerase chain reaction (RT-PCR).Transcription levels of SATB1,HoxA9,and EZH2 are normalized to GAPDH respectively.
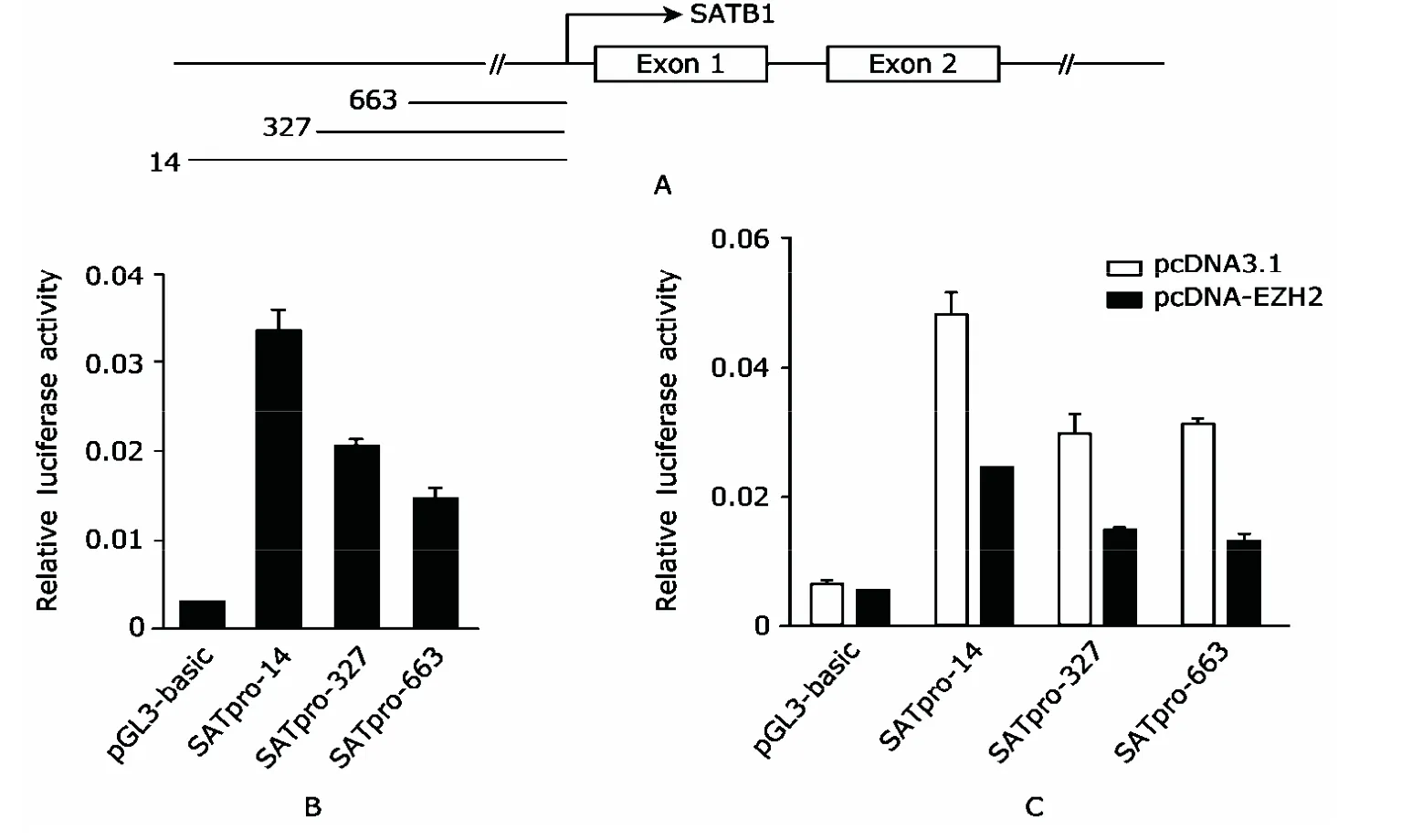
Figure 2.EZH2 represses promoter activity of SATB1 gene in HeLa epithelial cells.
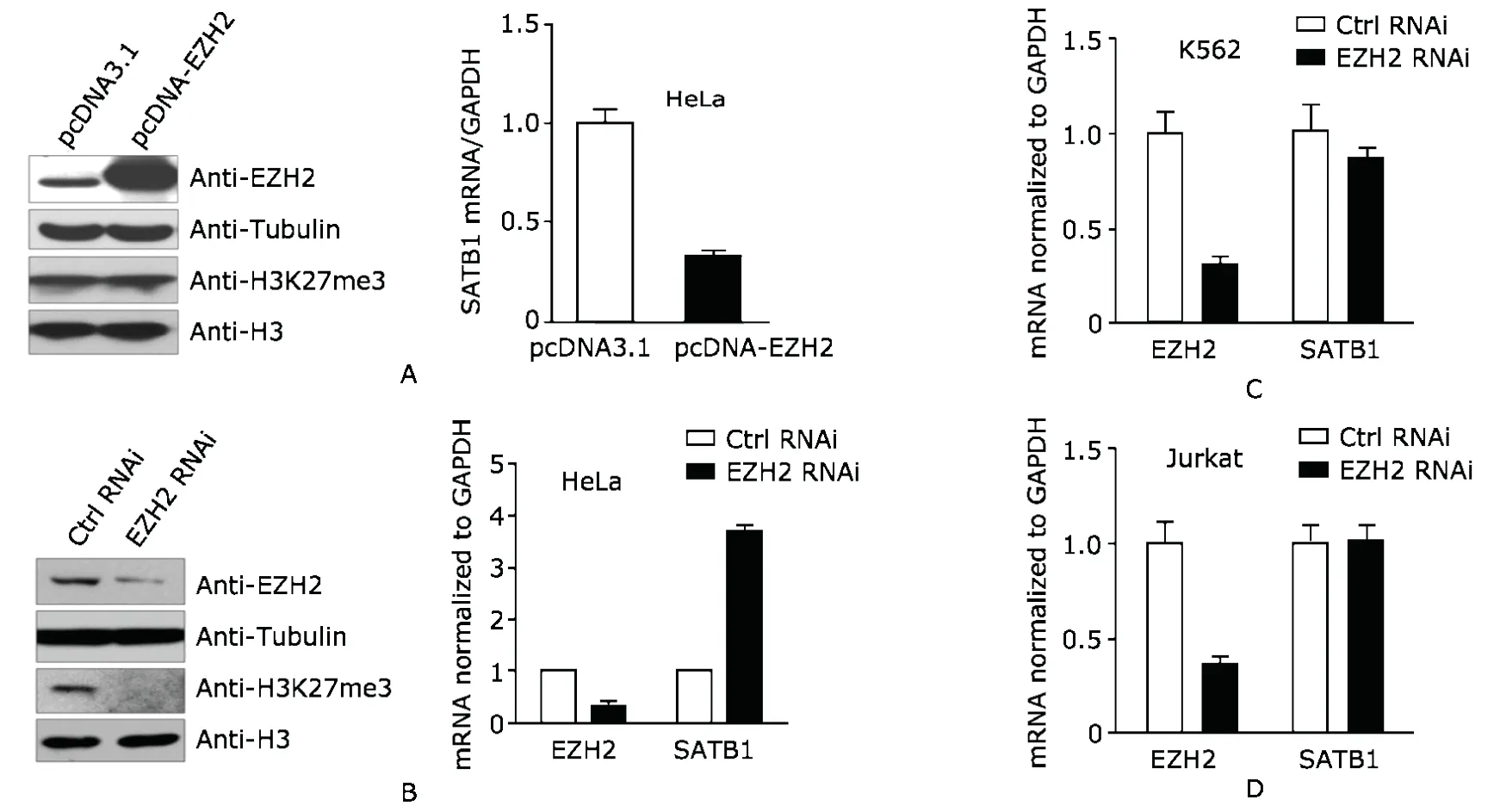
Figure 3.Expression of SATB1 is repressed by EZH2 in HeLa cells but not in K562 and Jurkat cells.Tubulin and total H3 serve as loading control in transfection,and the expressions measured by quantitative RT-PCR are normalized to GAPDH.
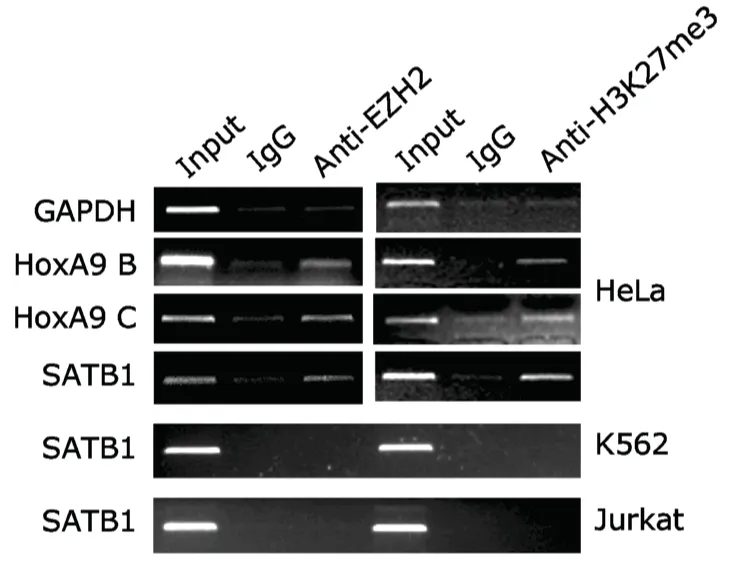
Figure 4.EZH2 and H3K27me3 are enriched within SATB1 proximal promoter in HeLa cells but not in K562 or Jurkat cells as demonstrated by chromatin immunoprecipitation (ChIP).IgG serves as a negative control to EZH2 and H3K27me3 antibodies.
DISCUSSION
Owing to the crucial regulatory roles of SATB1,10regulation of its own expression is important for understanding its biological functions.It is known that among adult tissues SATB1 expresses predominantly in thymus,and similar expression pattern was found between thymocytes and cells from other lineages.15,22Recent studies in T cells showed that upregulation of SATB1 by interleukin-4 in Th2 cells was mediated by STAT6,a transcription factor in the family of signal transducer and activator of transcription (STAT).26Bioinformatics analysis of SATB1 proximal promoter using UCSC Genome Browser also revealed conserved binding sites for transcription factors like BRN-2 and PAX5,27which are closely related to differentiation and proliferation of lymphocytes.Therefore,the relatively higher expression of SATB1 in T cells may be mediated by tissue-specific gene activators.
Mutation of SATB1 in mice results in deregulation of numerous genes and arrest of T cell development.10Likewise,aberrant over-expression of SATB1 in mammary cell lines was found to be able to reprogram gene expression,thereby promoting growth and metastasis of breast tumor.28However,data from Oncomine database showed that expression of SATB1 decreased significantly in many invasive ductal or lobular breast carcinoma samples compared with normal breast tissues.29In addition,a recent study using mammary cell lines revealed that increased expression of SATB1 did not promote breast cancer progression in the systems they studied.30The discrepancy suggests that well controlled low-level expression of SATB1 may be critical for normal cellular functions outside thymus.
However,the mechanism of SATB1 gene repression in cells other than T cells has not been clarified so far.In this study,we found that inhibition of DNA methylation and HDAC activity could induce SATB1 expression in HeLa epithelial cells,indicating SATB1 as a PRC2 target gene.Over-expression or knockdown of EZH2 in HeLa cells induced decrease or increase of SATB1 expression,respectively,further confirming the role of EZH2 in repression of SATB1,but the same repressive effect was not detected in K562 or Jurkat cells.These results suggest that SATB1 repression may be generally induced by PRC2 in cells other than T cells or erythroid cells.
In summary,our results revealed that PRC2 may contribute to repression of SATB1 gene in HeLa epithelial cells and maintenance of its weak expression.That repression was found to be likely mediated by recruitment of EZH2 to SATB1 proximal promoter region,leading to formation of H3K27me3 mark.In contrast,EZH2 did not attenuate the expression of SATB1 in K562 cells or Jurkat cells.Therefore,our findings provide at least one possible mechanism responsible for regulating SATB1 expression pattern,which may be helpful for further understanding of the roles of SATB1 in tumorigenesis of non-lymphoid and non-erythroid cells.
1.Cao R,Wang L,Wang H,et al.Role of histone H3 lysine 27 methylation inPolycomb-group silencing.Science 2002;298:1039-43.
2.Czermin B,Melfi R,McCabe D,et al.Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites.Cell 2002;111:185-96.
3.Kuzmichev A,Nishioka K,Erdjument-Bromage H,et al.Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein.Genes Dev 2002;16:2893-905.
4.Muller J,Hart CM,Francis NJ,et al.Histone methyltransferase activity of a Drosophila Polycomb group repressor complex.Cell 2002;111:197-208.
5.Sparmann A,van Lohuizen M.Polycomb silencers control cell fate,development and cancer.Nat Rev Cancer 2006;6:846-56.
6.Simon JA,Kingston RE.Mechanisms of polycomb gene silencing:knowns and unknowns.Nat Rev Mol Cell Biol 2009;10:697-708.
7.Boyer LA,Plath K,Zeitlinger J,et al.Polycomb complexes repress developmental regulators in murine embryonic stem cells.Nature 2006;441:349-53.
8.Bracken AP,Dietrich N,Pasini D,et al.Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions.Genes Dev 2006;20:1123-36.
9.Schwartz YB,Kahn TG,Nix DA,et al.Genome-wide analysis of Polycomb targets inDrosophila melanogaster.Nat Genet 2006;38:700-5.
10.Alvarez JD,Yasui DH,Niida H,et al.The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development.Genes Dev 2000;14:521-35.
11.Cai S,Han HJ,Kohwi-Shigematsu T.Tissue-specific nuclear architecture and gene expression regulated by SATB1.Nat Genet 2003;34:42-51.
12.Yasui D,Miyano M,Cai S,et al.SATB1 targets chromatin remodelling to regulate genes over long distances.Nature 2002;419:641-5.
13.Pavan Kumar P,Purbey PK,Sinha CK,et al.Phosphorylation of SATB1,a global gene regulator,acts as a molecular switch regulating its transcriptional activityin vivo.Mol Cell 2006;22:231-43.
14.Cai S,Lee CC,Kohwi-Shigematsu T.SATB1 packages densely looped,transcriptionally active chromatin for coordinated expression of cytokine genes.Nat Genet 2006;38:1278-88.
15.Kumar PP,Bischof O,Purbey PK,et al.Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus.Nat Cell Biol 2007;9:45-56.
16.Savarese F,Davila A,Nechanitzky R,et al.Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression.Genes Dev 2009;23:2625-38.
17.Wen J,Huang S,Rogers H,et al.SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression.Blood 2005;105:3330-9.
18.Gong H,Wang Z,Zhao GW,et al.SATB1 regulates beta-like globin genes through matrix related nuclear relocation of the cluster.Biochem Biophys Res Commun 2009;383:11-5.
19.Wang L,Di LJ,Lv X,et al.Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster.PLoS One 2009;4:e4629.
20.Lund R,Aittokallio T,Nevalainen O,et al.Identification of novel genes regulated by IL-12,IL-4,or TGF-β during the early polarization of CD4+lymphocytes.J Immunol 2003;171:5328-36.
21.Lund R,Ahlfors H,Kainonen E,et al.Identification of genes involved in the initiation of human Th1 or Th2 cell commitment.Eur J Immunol 2005;35:3307-19.
22.Dickinson LA,Joh T,Kohwi Y,et al.A tissue-specific MAR/SAR DNA-binding protein withunusual binding site recognition.Cell 1992;70:631-45.
23.van der Vlag J,Otte AP.Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation.Nat Genet 1999;23:474-8.
24.Hernández-Mu?oz I,Taghavi P,Kuijl C,et al.Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1.Mol Cell Biol 2005;25:11047-58.
25.Vire E,Brenner C,Deplus R,et al.The Polycomb group protein EZH2 directly controls DNA methylation.Nature 2006;439:871-4.
26.Ahlfors H,Limaye A,Elo LL,et al.SATB1 dictates expression of multiple genes including IL-5 involved in human Thelper cell differentiation.Blood 2010;116:1443-53.
27.Kent WJ,Sugnet CW,Furey TS,et al.The human genome browser at UCSC.Genome Res 2002;12:996-1006.
28.Han HJ,Russo J,Kohwi Y,et al.SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis.Nature 2008;452:187-93.
29.Rhodes DR,Kalyana-Sundaram S,Mahavisno V,et al.Oncomine 3.0:genes,pathways,and networks in a collection of 18,000 cancergene expression profiles.Neoplasia 2007;9:166-80.
30.Iorns E,Hnatyszyn HJ,Seo P,et al.The role of SATB1 in breast cancer pathogenesis.J Natl Cancer Inst 2010;102:1284-96.
 Chinese Medical Sciences Journal2010年4期
Chinese Medical Sciences Journal2010年4期
- Chinese Medical Sciences Journal的其它文章
- Endothelial Nitric Oxide Synthase Gene Polymorphisms Associated with Susceptibility to High Altitude Pulmonary Edema in Chinese Railway Construction Workers at Qinghai-Tibet over 4 500 Meters above Sea Level△
- Thoraco-abdominal Aorta Revascularization through a Retroperitoneal Approach
- Clinicopathological Features of Non-familial Colorectal Cancer with High-frequency Microsatellite Instability△
- NF-E2:a Novel Regulator of Alpha-hemoglobin Stabilizing Protein Gene Expression△
- Expression of Cyclooxygenase-2 and Its Relationship with Mismatch Repair and Microsatellite Instability in Hereditary Nonpolyposis Colorectal Cancer△
- Giant Hydroureteronephrosis Associated with Ipsilateral Inguinal Hernia and Contralateral Hydronephrosis:a Case Study
