Inhibition of SIRT1 Increases EZH2 Protein Level and Enhances the Repression of EZH2 on Target Gene Expression△
Lu Lu,Lei Li,Xiang Lü,Xue-song Wu,De-pei Liu*,and Chih-chuan Liang
National Laboratory of Medical Molecular Biology,Institute of Basic Medical Sciences,Chinese Academy of Medical Sciences &Peking Union Medical College,Beijing 100005,China
POLYCOMB group (PcG) proteins were first shown to play essential roles in regulating body formation ofDrosophila melanogaster,which needs the correct spatial expression of HOX genes.1PcG proteins form multi-protein complexes and exert repressive effects on numerous genes controlling different pathways involved in development,differentiation,and cell fate decisions.2-4PcG proteins fall into two classical types,namely Polycomb repressive complexes 1 and 2 (PRC1 and PRC2),and PRC2 could catalyze trimethylation of nucleosomal histone H3 lysine 27 (H3K27) with its methyltransferase activity,forming trimethylated H3K27 (H3K27-me3) to initiate gene silencing.5-8PRC2 is comprised of enhancer of Zeste homolog 2 (EZH2),SUZ12,and EED,among which EZH2 is an enzymatic subunit,and the other two are required for the enzyme activity.9
EZH2 expression level is high in stem cells and decreases greatly in differentiated tissues,but recent studies on human tumors showed that it is frequently over-expressed in malignant tumors and associated with poor prognosis.10,11It is thus important to understand the mechanisms regulating the expression level of EZH2.Bracken et al12reported that EZH2 was downstream of the pRB/E2F pathway,which is an essential pathway in cell proliferation and tumorigenesis,and was also amplified in invasive tumors.A study showed that androgens can suppress EZH2 expressionviaretinoblastoma and p130-dependent pathways in some prostate cancer cells.13It was also shown that some microRNAs,such as miR-101,miR-26a,miR-214,and miR-138,contributed to regulation of EZH2 level at the post-transcriptional level.14-17
In addition to those internal cellular regulators,EZH2 level is also affected by exogenous drugs.For example,the S-adenosylhomocysteine hydrolase inhibitor 3-deazaneplanocin A was shown to effectively reduce cellular levels of the 3 components of PRC2 and inhibit associated H3K27 methylation in cancer cells.18Another study found that dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells.19Treatment with histone deacetylase inhibitor LBH589 or LAQ824 in human leukemia cells also depleted the protein levels of EZH2,SUZ12,and EED.20
SIRT1,a class III histone deacetylase,has been shown to regulate many other nuclear regulators,and play roles in a variety of cellular processes.21SIRT1 was also found to have protein-protein interactions with EZH2,and forms the PRC4 complex in certain conditions.22That study indicated close relations between SIRT1 and EZH2,but it is not clear whether SIRT1 contributes to regulation of EZH2 expression.In this study,we explored the potential regulatory role of SIRT1 in the expression of EZH2 and its further effects on EZH2 repression of SATB1,which was identified as abona fideEZH2 target gene in our previous study.23
MATERIALS AND METHODS
Cell culture and establishment of stable SIRT1 RNAi cell lines
HeLa cells (Peking Union Medical College Culture Collection)and GP-293 cells (Takara Bio,Ostu,Shiga,Japan) were maintained in Delbecco’s modified Eagle’s medium (Invitrogen,Carlsbad,CA,USA) supplemented with 10% fetal bovine serum.
The targeting sequence (passenger strand) for SIRT1 knockdown is 5’-GATGAAGTTGACCTCCTCA-3’.The shRNA oligos were ligated into pSirenRetroQ (Takara Bio) atBamHIandEcoRIsites to generate pSiren-shSIRT1.The construct pSiren-shLuc expressing shRNA against luciferase was used as control.The two types of plasmids were separately transfected into GP-293 cells along with pVSV-G plasmid (Takara Bio) to generate retroviruses.Sixty hours after transfection,the media were collected,filtered,and stored at-80°C for later infections of HeLa cells.Forty-eight hours after infection,the HeLa cells were added with 1 μg/mL puromycin and maintained for 7-10 days to generate stable knockdown cell lines (SIRT1 RNAi and Control RNAi).
Extraction of cellular proteins and Western blot
To generate whole cell lysate,the SIRT1 RNAi and Control RNAi HeLa cells were lysed with RIPA buffer [50 mmol/L Tris-HCl (pH 8.0),150 mmol/L NaCl,1% NP-40,0.5%sodium deoxycholate,0.1% sodium dodecyl sulfate (SDS),and protease inhibitor cocktail from Sigma-Aldrich (St.Louis,MO,USA)]and the extracts were sonicated for 15 seconds with a Branson 150 sonicator (Branson Ultrasonics,Danbury,CT,USA).
EZH2 proteins mainly concentrate in the chromatin,and a minor fraction can also be detected in the cytoplasm.To further investigate which part is responsible for the possible change in protein level,we used fractional extraction method to detect different parts of the EZH2 protein pool.For fractional extraction,6×106SIRT1 RNAi and Control RNAi HeLa cells were separately suspended in 400 μL solution A [10 mmol/L HEPES (pH 7.9),10 mmol/L KCl,1.5 mmol/L MgCl2,0.34 mol/L sucrose,10% glycerol,1 mmol/L dithiothreitol (DTT),and protease inhibitor cocktail from Sigma-Aldrich].Triton X-100 (Sigma-Aldrich)was added to the cell suspension and the final concentration was 0.1%.After incubation on ice for 5 minutes followed by centrifugation for 4 minutes (1300 ×g),the supernatant was harvested as cytoplasmic fraction.The nuclei were washed with solution A,lysed in 400 μL solution B[3 mmol/L ethylene diamine tetraacetic acid (EDTA),0.2 mmol/L ethylene glycol tetraacetic acid,1 mmol/L DTT,and protease inhibitor cocktail from Sigma-Aldrich],and incubated on ice for 10 minutes.After centrifugation at 1700 ×gfor 4 minutes,the isolated chromatin pellet was washed with solution B,centrifuged at 10 000 ×gfor 1 minute,re-suspended in 400 μL 1× sample loading buffer(Beyotime,Haimen,Jiangsu,China),and sonicated for 30 seconds (Branson 150 sonicator,Branson Ultrasonics),producing the chromatin fraction.
The protein samples were then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDSPAGE) and transferred to membranes.The blots were probed by anti-EZH2 (Cell Signaling Technology,Boston,MA,USA),anti-SIRT1 (Santa Cruz Biotechnology,Santa Cruz,CA,USA),anti-H3K27me3 (Millipore,Billerica,MA,USA),anti-H3 (Abcam,Cambridge,MA,USA),and antitubulin (Abcam) antibodies.Apart from being loading controls,the H3 level and tubulin level serve as the fractional maker for chromatin and cytoplasmic fractions respectively.
Reverse transcription-polymerase chain reaction(RT-PCR) and real-time RT-PCR
The protein level changes detected by Western blot may result from the changes of gene transcription,thus RT-PCR was perfomred to detect the possible transcriptional changes.Total RNA from SIRT1 RNAi and Control RNAi HeLa cells were isolated with TRIzol reagent (Invitrogen)and reverse transcription was performed following the manufacturer’s manual (New England Biolabs,Ipswich,MA,USA).For the classical RT-PCR,the PCR products were separated by 2% agarose gel electrophoresis.The gel was then stained with distilled water containing 0.2 μg/mL ethidium bromide followed by destai-ning with distilled water for 10 minutes.The gel was then scanned with UV imaging system (Bio-Rad,Hercules,CA,USA).Real-time PCR was performed using SYBR Premix Ex Taq (Takara Bio)on Bio-Rad’s iQ5 real-time thermal cycler (Bio-Rad).Primer sequences are as follows:EZH2,5’-GCCAGACTGGGAAGAAATCTG-3’ (forward),5’-TGTGTTGGAAAATCCAAGTCA-3’ (reverse);SATB1,5’-GATCTATGAATAAGCCTTTGGAG-3’ (forward),5’-TTTCGTCCTGGTATATTCGGT-3’(reverse);SIRT1,5’-GATTGGCACAGATCCTCGAAC-3’(forward),5’-CCCCAGCTCCAGTTAGAACTA-3’ (reverse);GAPDH,5’-GGTGAAGGTCGGAGTCAACGGA-3’(forward),5’-GAGGGATCTCGCTCCTGGAAGA-3’(reverse) (internal control).
Protein stability assay
In addition to the changes in transcription,those in protein stability can also alter levels of specific proteins,thus the protein stability assay was applied.To determine the half-life of EZH2 proteins,HeLa cells were treated with 0.1 μg/mL of cycloheximide (CHX) (Sigma-Aldrich) for different time (0 hour,2 hours,4 hours,6 hours,and 8 hours),and the protein level of EZH2 was then measured by Western blot.Based on the results observed in that experiment,three time points were selected for the treatment with 0.1 μg/mL of CHX in the SIRT1 RNAi and Control RNAi cells.Then the treated cells were washed three times with cold phosphate-buffered saline to remove CHX,followed by extraction of whole cell lysate and Western blot assay.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as described in the Upstate protocol (Millipore) with a few minor modifications.After crosslinking with 1% formaldehyde,the isolated nuclei from SIRT1 RNAi and Control RNAi HeLa cells were lysed in SDS lysis buffer [1% SDS,10 mmol/L EDTA,and 50 mmol/L Tris-HCl (pH 8.1)]and sonicated with a Branson 150 sonicator (Branson Ultrasonics) to shear the fragments down to 200-750 bp.Antibodies against EZH2 (Cell Signaling Technology),H3K27me3 (Millipore),and SIRT1(Santa Cruz Biotechnology) were used for immunoprecipitation.The ChIP products were then detected by PCR,followed by agarose gel electrophoresis and staining as described above.The primers used in this procedure are as follows:EZH2-promoter,5’-GGGCCAAATAAAAGCGATGG-3’ (forward),5’-CTGCCTTCTGAGTCCCACCG-3’ (reverse);SIRT1-promoter,5’-AAAGTCACGCAGGTAATTGATGC-3’ (forward),5’-GAGATAACCAGATGTAAAACGAGGG-3’ (reverse);SATB1-promoter,5’-ACGTCGTTTCCCCAGCAC-3’ (forward),5’-AAACGTCTAGAAGAGTAGCCATGAGAA-3’ (reverse);GAPDH-promoter,5’-TACTAGCGGTTTTACGGGCG-3’ (forward),5’-TCGAACAGGAGGAGCAGAGAG-3’ (reverse) (negative control)
RESULTS
Protein level of EZH2 upon SIRT1 depletion
Western blot results showed efficient SIRT1 knockdown in the SIRT1 RNAi cells.The protein level of EZH2 in whole cell lysate increased upon SIRT1 depletion (Fig.1A).As EZH2 is the major enzyme for H3K27 methylation,the H3K27me3 level was detected,and the results showed that the increase of EZH2 does not cause global changes of H3K27me3 level (Fig.1B).The results of fractional extraction showed that in the cytoplasmic fraction,EZH2 protein level remained unchanged upon SIRT1 knockdown(Fig.1C).In the chromatin fraction,in contrast,EZH2 protein level was found elevated upon SIRT1 knockdown,suggesting that the EZH2 recruited to chromatin may be the source of detected increase in EZH2 protein expression(Fig.1D).In the chromatin fraction,histone H3 shows strong signals in Western blot,while tubulin,which is a cytoplasmic marker,was barely detected,confirming the reliability of the Western blot results.
Transcription of EZH2 upon SIRT1 depletion
The increased protein level of EZH2 may result from either or both increased transcription and increased protein stability.The results of RT-PCR analysis showed that the mRNA level of EZH2 remained unchanged in SIRT1 RNAi and Control RNAi cells (Fig.2A).ChIP assay showed strong enrichment of SIRT1 at its own promoter,which decreased upon SIRT1 depletion (Fig.2B).In contrast,no SIRT1 binding was detected at EZH2 promoter,similar with the result at GAPDH promoter,the negative control (Fig.2B).Taken together,SIRT1 may not be recruited to the EZH2 promoter to regulate its transcription.
Effect of SIRT1 depletion on protein stability of EZH2
We examined if SIRT1 depletion affects the protein stability of EZH2.After treating HeLa cells with 0.1 μg/mL CHX for different time periods,we noticed that EZH2 protein level was stable within four hours but apparently reduced after six hours (Fig.3A).Based on this finding,three time points(0 hour,4 hour,and 8 hour) were selected for detection of the protein stability of EZH2 in SIRT1 RNAi and Control RNAi cells.The Western blot results showed similar decrease rate of EZH2 protein level between the two cell lines within four hours,but the decline became slower in SIRT1 RNAi cells from 4 hour to 8 hour (Fig.3B).These results suggest that upon SIRT1 depletion,the protein stability of EZH2 increases,contributing to the elevation of its protein level.
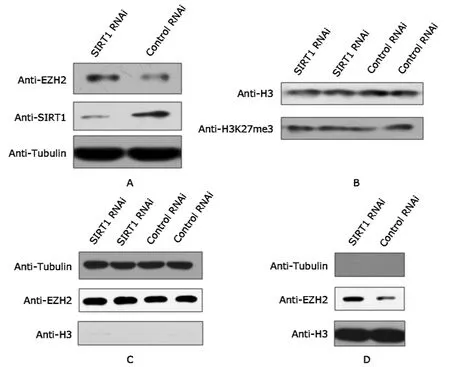
Figure 1.Western blot results of EZH2 protein level upon SIRT1 knockdown.
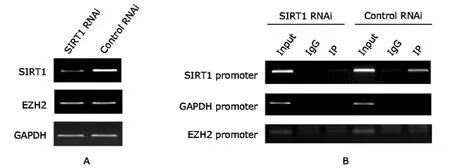
Figure 2.Reverse transcription-polymerase chain reaction (RT-PCR) results of EZH2 mRNA level and chromatin immunoprecipitation(ChIP) results of SIRT1 binding at EZH2 promoter.
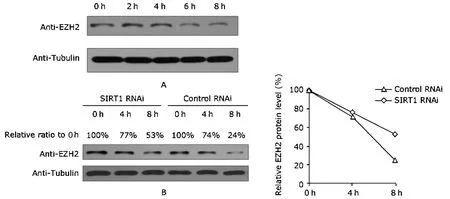
Figure 3.Effects of SIRT1 knockdown on the protein stability of EZH2 as detected with Western blot.
Enhanced suppression of the target gene of EZH2
We detected the expression of SATB1,abona fideEZH2 target gene,in SIRT1 RNAi and Control RNAi cells.The realtime RT-PCR results showed that SATB1 mRNA decreased in response to SIRT1 knockdown (Fig.4A).The amplification products of real-time PCR were confirmed by agarose gel electrophoresis and ethidium bromide staining (Fig.4A).
EZH2 ChIP showed that in SIRT1 RNAi cells,the enrichment of EZH2 at SATB1 promoter is higher than that in Control RNAi cells,while no enrichment of EZH2 was detected at GAPDH promoter,the negative control (Fig.4B).Since repression by EZH2 is initiated by trimethylation of histone H3K27,H3K27me3 was also detected at SATB1 promoter.The results showed that H3K27me3 level increased upon SIRT1 depletion (Fig.4B).
SIRT1 ChIP results showed no enrichment of SIRT1 at SATB1 promoter region,but strong enrichment at the promoter of its own gene locus,which dramatically decreased upon SIRT1 depletion (Fig.4B).As the negative control,GAPDH promoter region demonstrated no SIRT1 enrichment (Fig.4B).
Taken together,these results indicate that the repression of SATB1,a target gene of EZH2,may be strengthened by SIRT1 depletion.

Figure 4.Enhanced suppression of EZH2 target gene SATB1 upon SIRT1 depletion.
DISCUSSION
SIRT1 has been shown to bind with and deacetylate many transcription factors and other nuclear factors,such as p53,FOXO3a,E2F1,HIC1,and Suv39H1.24-28Through those interactions,SIRT1 functions in cell survival,metabolism,DNA repair,and tumorigenesis.In this study,we found that SIRT1 depletion leads to increased protein level of EZH2 in HeLa cells.It is known that the pRB-E2F pathway is the major regulatory route controlling EZH2 expression,12and SIRT1 was reported to be associated with E2F1,27suggesting that SIRT1 might regulate EZH2 expression at transcriptional level.Nevertheless,the results of the present study showed that the mRNA level of EZH2 was not affected by SIRT1 knockdown,and no SIRT1 enrichment was detected at EZH2 promoter.On the other hand,the protein stability assay showed increased EZH2 protein stability upon SIRT1 knockdown.Those findings suggest that SIRT1 might participate in regulation of EZH2 expression at the protein level,by affecting the protein stability.
Increased EZH2 protein level upon SIRT1 knockdown then leads to reduced expression of SATB1,which was identified previously as abona fideEZH2 target gene in HeLa cells.23It is still important to investigate if SIRT1 directly regulates SATB1 expression in HeLa cells,because SIRT1 can deacetylate several histone substrates,such as H4K16Ac,the modification status of which has been studied to be associated with gene transcription.29,30SIRT1 has also been shown to be present at the promoters of some tumor suppressor genes,and its deficiency results in a hyperacetylation of H4K16,which in turn induces expression of those genes.31The ChIP assay showed no SIRT1 binding at SATB1 promoter,suggesting that SIRT1 affects the expression of this PcG target gene in an H4K16Ac-independent manner.
Previous studies have shown that SIRT1 played many roles in different cellular activities.Regarding cancer,the genetic evidence from a recent study using SIRT1 knockout mice and human cancer samples supports the tumor suppressor role of SIRT1.32,33Our results showed that knockdown of SIRT1 elevated the protein level of EZH2,which is abona fideoncogene,further confirming SIRT1’s tumor suppressor role.Studies on the regulation mechanism may provide important information for understanding of the roles of SIRT1 and EZH2 in the tumorigenesis of certain cancers.
1.Kennison JA.The Polycomb and trithorax group proteins of Drosophila:trans-regulators of homeotic gene function.Annu Rev Genet 1995;29:289-303.
2.Bracken AP,Dietrich N,Pasini D,et al.Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions.Genes Dev 2006;20:1123-36.
3.Schwartz YB,Kahn TG,Nix DA,et al.Genome-wide analysis of Polycomb targets in Drosophila melanogaster.Nat Genet 2006;38:700-5.
4.Boyer LA,Plath K,Zeitlinger J,et al.Polycomb complexes repress developmental regulators in murine embryonic stem cells.Nature 2006;441:349-53.
5.Czermin B,Melfi R,McCabe D,et al.Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites.Cell 2002;111:185-96.
6.Müller J,Hart CM,Francis NJ,et al.Histone methyltransferase activity of a Drosophila Polycomb group repressor complex.Cell 2002;111:197-208.
7.Kuzmichev A,Nishioka K,Erdjument-Bromage H,et al.Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein.Genes Dev 2002;16:2893-905.
8.Cao R,Wang L,Wang H,et al.Role of histone H3 lysine 27 methylation in Polycomb-group silencing.Science 2002;298:1039-43.
9.Pasini D,Bracken AP,Jensen MR,et al.Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity.EMBO J 2004;23:4061-71.
10.Simon JA,Lange CA.Roles of the EZH2 histone methyltransferase in cancer epigenetics.Mutat Res 2008;647:21-9.
11.Mills AA.Throwing the cancer switch:reciprocal roles of polycomb and trithorax proteins.Nat Rev Cancer 2010;10:669-82.
12.Bracken AP,Pasini D,Capra M,et al.EZH2is downstream of the pRB-E2F pathway,essential for proliferation and amplified in cancer.EMBO J 2003;22:5323-35.
13.Bohrer LR,Chen S,Hallstrom TC,et al.Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways:a potential mechanism of androgen-refractory progression of prostate cancer.Endocrinology 2010;151:5136-45.
14.Varambally S,Cao Q,Mani RS,et al.Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer.Science 2008;322:1695-9.
15.Ciarapica R,Russo G,Verginelli F,et al.Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma.Cell Cycle 2009;8:172-5.
16.Juan AH,Kumar RM,Marx JG,et al.Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells.Mol Cell 2009;36:61-74.
17.Kisliouk T,Yosefi S,Meiri N.MiR-138 inhibits EZH2 methyltransferase expression and methylation of histone H3 at lysine 27,and affects thermotolerance acquisition.Eur J Neurosci 2011;33:224-35.
18.Tan J,Yang X,Zhuang L,et al.Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells.Genes Dev 2007;21:1050-63.
19.Dimri M,Bommi PV,Sahasrabuddhem AA,et al.Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells.Carcinogenesis 2010;31:489-95.
20.Fiskus W,Pranpat M,Balasis M,et al.Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells.Mol Cancer Ther 2006;5:3096-104.
21.Haigis MC,Sinclair DA.Mammalian sirtuins:biological insights and disease relevance.Annu Rev Pathol 2010;5:253-95.
22.Kuzmichev A,Margueron R,Vaquero A,et al.Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation.Proc Natl Acad Sci U S A 2005;102:1859-64.
23.Li L,Lu L,Lü X,et al.Epigenetic repression of SATB1 by Polycomb group protein EZH2 in epithelial cells.Chin Med Sci J 2010;25:199-205.
24.Vaziri H,Dessain SK,Ng Eaton E,et al.hSIR2(SIRT1)functions as an NAD-dependent p53 deacetylase.Cell 2001;107:149-59.
25.Brunet A,Sweeney LB,Sturgill JF,et al.Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase.Science 2004;303:2011-5.
26.Chen WY,Wang DH,Yen RC,et al.Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses.Cell 2005;123:437-48.
27.Wang C,Chen L,Hou X,et al.Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage.Nat Cell Biol 2006;8:1025-31.
28.Vaquero A,Scher M,Erdjument-Bromage H,et al.SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation.Nature 2007;450:440-4.
29.Vaquero A,Scher M,Lee D,et al.Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin.Mol Cell 2004;16:93-105.
30.Kurdistani SK,Tavazoie S,Grunstein M.Mapping global histone acetylation patterns to gene expression.Cell 2004;117:721-33.
31.Pruitt K,Zinn RL,Ohm JE,et al.Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation.PLoS Genet 2006;2:e40.
32.Wang RH,Sengupta K,Li C,et al.Impaired DNA damage response,genome instability,and tumorigenesis in SIRT1 mutant mice.Cancer Cell 2008;14:312-23.
33.Deng CX.SIRT1,is it a tumor promoter or tumor suppressor? Int J Biol Sci 2009;5:147-52.
 Chinese Medical Sciences Journal2011年2期
Chinese Medical Sciences Journal2011年2期
- Chinese Medical Sciences Journal的其它文章
- Risk Factors Analysis on Traumatic Brain Injury Prognosis
- Erythropoietin Receptor Positive Circulating Progenitor Cells and Endothelial Progenitor Cells in Patients with Different Stages of Diabetic Retinopathy△
- Immediate Surgical Intervention for Penile Fracture:a Case Report and Literature Review
- Clinical Treatment and Anatomy Study of Maxillary First Molars with Five Root Canals
- Magnetic Resonance Urography and X-ray Urography Findings of Congenital Megaureter
- Changes of Nerve Growth Factor in Amniotic Fluid and Correlation with Ventriculomegaly△
