Proper progression of neurogenesis relies on a defined pattern of SUMOmodified proteins
Mario García-Domínguez
Neurogenesis is a complex process involving the orchestration of many transcription factors and other proteins. Fine regulation of their activities is crucial for proper progression of neurogenesis. A few decades ago, covalent attachment of Small Ubiquitin-like MOdifier(SUMO) to other proteins was revealed as a major regulator of protein activities,constituting an essential posttranslational modification system in vertebrates. Since then, hundreds of proteins have been shown to be targets of SUMO, which is implicated in controlling many relevant processes in eukaryotic cells. These include the development and function of the nervous system, with SUMO tightly linked to synapsis and to neurodegenerative diseases (Yau et al., 2020).The SUMO protease SENP7 has been involved in neurogenesis (Juárez-Vicente et al., 2016),while sumoylation of BRAF35, a subunit of the LSD1 histone demethylase complex, assures the undifferentiated state of neural progenitors(Ceballos-Chávez et al., 2012). In addition,increased sumoylation in mouse brain-derived neural stem cells results in enhanced neuronal differentiation (Bernstock et al., 2019).However, in general, the implication of SUMO in neuronal differentiation and nervous system development has been poorly studied. We have recently developed a proteomic study aimed at identifying SUMO targets associated either with proliferation or with neuronal differentiation conditions, which has highlighted the relevance of SUMO modification for proper progression of neurogenesis. Here, we discuss our recent results, present specific SUMO targets that are key to the process, and indicate future research directions to uncover the molecular mechanisms underlying SUMO modification of relevant neurogenesis-associated factors.
SUMO is a small polypeptide of approximately 100 amino acids, similar to ubiquitin, that is able to covalently attach to other proteins through the ε-amino group of a lysine (K)residue to modulate its function/activity(Flotho and Melchior, 2013). Target K is often included in the consensus ψKxE (ψ, large hydrophobic residue; x, any residue). Three major functional species of SUMO have been described in vertebrates, SUMO1–3. While most SUMO1 in the cell appears conjugated to proteins, a good fraction of the closely related paralogs SUMO2 and SUMO3 (SUMO2/3) pool is free to be rapidly conjugated to proteins in response to a variety of stress conditions. In addition to specific activating (SAE1-UBA2) and conjugating (UBC9) enzymes, SUMO ligases and proteases participate in SUMO attachment and detachment from targets, respectively, making the modification a regulated and reversible process.
The involvement of SUMO in regulating most relevant cellular processes anticipates its participation in the control of neurogenesis. To shed light on this aspect, we recently developed a SILAC-based proteomic approach to compare the SUMO proteome of proliferating and neuronal differentiating pluripotent embryonic carcinoma P19 cells (Correa-Vázquez et al.,2021). From this analysis, we identified up to 318 differentially sumoylated proteins between the two conditions. They included proteins that were similarly expressed under both conditions but sumoylated in one of them, as proteins expressed in one of the conditions in which they were sumoylated. Thus, we uncovered distinct patterns of sumoylated proteins associated with proliferation and neuronal differentiation (Figure 1A). For a selection of proteins, we made use of sumoylation mutants in gain-of-function experiments to gain insight into the role the modification plays during neurogenesis. Mutants consisted of substitution of target K residues by arginine(KR). We analyzed the effects of wild-type (WT)and mutant proteins in cellular and embryonic models of neurogenesis in which we induced neurogenesis by the neurogenic factors NeuroD2 and Neurogenin2, respectively. In all cases, the effects of mutants significantly differed from those of WT proteins. For most proteins, sumoylation was associated with proper progression of neurogenesis (Figure 1A). This was, for instance, the case of Prospero homeobox 1, essentially expressed and sumoylated under differentiation conditions.SUMO was necessary for its previously reported function in cell cycle withdrawal of neural progenitors (Misra et al., 2008), indicating that sumoylation of relevant differentiationassociated factors is required for neurogenesis.Interestingly, although the pluripotencyassociated factors Spalt-like transcription factor 4 and undifferentiated embryonic cell transcription factor 1 (UTF1) (Rao et al.,2010; Raina et al., 2021) were expressed and sumoylated under proliferation conditions, their modification was also required for neuronal differentiation. This illustrates how sumoylation of proliferation-associated factors is also key to neurogenesis. In turn, tripartite motif containing 24 expression remained unchanged between conditions, and sumoylation under differentiation was required for neurogenesis.Thus, regardless of whether these proteins are expressed under one or the other condition,SUMO-mediated modification of them is associated with proper neurogenesis.
We also studied potassium channel tetramerization domain containing 15 (KCTD15)(Correa-Vázquez et al., 2021). This protein was expressed and sumoylated under differentiation conditions, and unexpectedly, overexpression of the WT protein was associated with impaired neurogenesis. Thus, the effect on neurogenesis of KCTD15 sumoylation was opposite to that of other analyzed proteins, suggesting that KCTD15 slows neurogenesis in a SUMOdependent manner for accurate timing and successful accomplishment of the process.Moreover, the KR mutant unusually displayed neurogenic activity in the developing neural tube in the absence of induced neurogenesis.This is a typical feature of neurogenic factors such as Neurogenin2 or NeuroD2, which is not expected for mutant KCTD15. As mentioned,Prospero homeobox 1 expression was able to stimulate neurogenesis, but only under neuronal differentiation conditions induced by neurogenic factors. We do not know the mechanisms underlying this KCTD15-KR effect,but it might be an interesting aspect to study and perhaps to exploit.
However, among the identified proteins,UTF1 aroused the greatest interest. It was not previously described as a SUMO target, and detailed characterization demonstrated SUMOdependent regulation of its transcriptional activity at several levels. An important feature of embryonic stem cells is the poised state of many relevant developmentally regulated genes, designated bivalent genes. They harbor either active (trimethylated lysine 4 of histone 3) or repressive (trimethylated lysine 27 of histone 3) chromatin marks,with polycomb-repressive complex 2 (PRC2)being involved in establishing the repressive mark. It was previously reported that UTF1 controls the expression of bivalent genes by two mechanisms (Jia et al., 2012). On one hand, it limits the localization of PRC2 to regulated promoters, avoiding deep repression of bivalent genes. On the other hand, UTF1 also participates in recruiting the decapping mRNA 1A (DCP1A) component of the mRNAdecapping complex for degradation of leakage mRNAs. This dual mechanism ensures that the expression of bivalent genes is kept to a minimum but prone to activation. Notably, we have described that SUMO attachment to UTF1 modulates both mechanisms (Correa-Vázquez et al., 2021) (Figure 1B). First, sumoylation alters UTF1 affinity for chromatin, with better attachment in the unmodified state. Thus,enhanced chromatin association of the KR mutant, behaving as a dominant negative molecule, may result in increased displacement of factors/complexes from chromatin. Second,DCP1A recruitment appeared to occur in a SUMO-dependent manner. Intriguingly, by analyzing the effects of WT and KR UTF1 on a battery of bivalent genes, we observed a set of genes negatively affected by the WT protein,while another set was negatively affected by the KR version. Interestingly, this correlated with the previously reported effect ofUtf1knockout in mouse embryonic stem cells (Jia et al., 2012). Those genes negatively affected by the WT protein were genes reported to be upregulated upon knockout, while those negatively affected by the KR mutant were described as downregulated in knockout cells.Therefore, we can conclude that both the absence of UTF1 and defective sumoylation lead to similar altered gene expression.Differences observed between genes probably depend on the chromatin context. The presence or absence of additional gene-specific factors and/or chromatin complexes should explain why some genes are upregulated while others are downregulated in the absence of functional UTF1. In relation to this, we can speculate that UTF1-KR interference with recruitment of other factors/complexes affects not only the PRC2 complex but also other factors/complexes,including transcription activators. Moreover,we cannot exclude the possibility that UTF1-KR also affects the recruitment of differentiationassociated factors/complexes just mobilized upon induction of differentiation and needed for transcriptional control of bivalent genes. For instance, in the case of those genes negatively affected by the KR mutant, two possible and not mutually excluding scenarios can be predicted: i) UTF1-KR may interfere with de novo recruitment of differentiation-associated factors/complexes for gene activation upon induction of differentiation. ii) UTF1-KR may interfere in the proliferative state with activators contributing to the poised state of genes, leading to a more repressive chromatin conformation and thereby decreasing activation upon induction of differentiation.Our proteomic analysis revealed that UTF1 is strongly downregulated after four days of differentiation, but the rate of desumylation was greater than the rate of protein decrease.A possibility is that desumylation leads to protein degradation, since SUMO attachment has been proposed to protect against ubiquitin/proteasome-mediated degradation in certain cases (Ulrich, 2005). This would mean that nonsumoylated UTF1 would have limited opportunity to interfere with the recruitment of other factors/complexes. We did not observe differences in stabilities between WT and KR proteins at short times. However, even if this is the case, faint and transitory interference at very early stages of differentiation, when desumoyed UTF1 is still abundant, might have a great impact on neurogenesis at later stages.It would be of interest to establish the kinetics of UTF1 desumylation and downregulation following the induction of neuronal differentiation. UTF1 is a key transcription factor associated with stemness and development in placental mammals and is also implicated in cancer (Raina et al., 2021). Thus,detailed dissection of its operating mechanisms is of high interest.
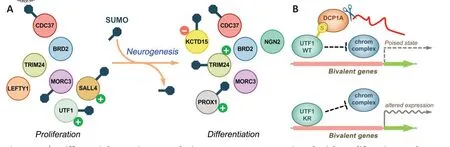
Figure 1 |Differential protein sumoylation patterns are associated with proliferation and neuronal differentiation.(A) A SILAC approach permitted the identification of protein sumoylation patterns of proliferating and neuronal differentiating cells. Identification of the SUMO proteome during neurogenesis indicates different subsets of sumoylated proteins under proliferation and neuronal differentiation conditions. SILAC differences were explained either by proteins equally expressed under both conditions but sumoylated in one of them, as by proteins expressed in one condition in which they are sumoylated. The study of selected proteins indicated that sumoylation may result in both positive (+) and negative (–) effects on neurogenesis. (B) UTF1 stands out among the key SUMO targets whose modification participates in the progression of neurogenesis. Sumoylation (S) of WT UTF1, by modulating UTF1 affinity for chromatin,limits to some extent access of chromatin regulatory complexes (chrom complex) to regulated promoters at the same time that it recruits DCP1A for degradation of leaking mRNAs, resulting in a poised expression state of bivalent genes. Nonsumoylable (KR) mutant UTF1 displays enhanced localization to promoters and fails to recruit DCP1A, leading to altered gene expression. NGN2: Neurogenin 2.
Undoubtedly, as occurs in the case of many other relevant cellular processes, sumoylation is critical for neurogenesis. Many of the players of the process use SUMO to modulate their activities. Thus, depending on the target,selective abolition of SUMO modification may lead to opposite effects. In turn, global alteration of sumoylation should lead to unique general effects. In relation to this,we have previously shown that enhancing global sumoylation in neurodevelopmental models, either by overexpressing different SUMO species or by knocking down the SUMO protease SENP7, leads to impaired neurogenesis(Juárez-Vicente et al., 2016). Conversely, it has been reported that UBC9 overexpression in adult mouse-derived neural stem cells leads to increased global sumoylation and enhanced neuronal differentiation (Bernstock et al.,2019). This might suggest that the association of SUMO with a neural undifferentiated state would be restricted to development. However,far from this interpretation, it probably means that establishment of highly specialized differentiation programs in stem cells requires fine tuning of a number of specific factors and that minor alteration of the activity of any of them suffices to abrogate neurogenesis.In other words, if just one key factor whose activity depends on sumoylation fails to be modified, underlying control systems should assure interruption of such a complex process.Therefore, it appears critical to precisely study the consequences of blocking sumoylation of each isolated key factor. In this context, it would be desirable in upcoming research to approach this topic by substitution of endogenous genes by their corresponding sumoylation mutant versions, for instance, by making use of CRISPR/Cas9 technology. However, discrete mutations impairing sumoylation may not result in dramatic phenotypes. Although milder effects at early developmental stages do not exclude severe consequences at later stages,in some cases, this probably results from additional overlapping control levels of protein activities. Nonetheless, a fundamental role of SUMO relies on the maintenance of complex stability, especially in the case of chromatinassociated complexes (García-Domínguez and Reyes, 2009). Thus, compared with moderate outcomes of discrete mutations, more severe phenotypes would be expected from simultaneous mutation of several proteins in a complex or pathway.
Research at the MGD lab is currently supported by grants PGC2018-094232-B-I00 from the Ministry of Science, Innovation and Universities (MICIU), Spain, and CV20-93141 from the Andalusian regional government(both co-financed by the European Regional Development Fund). We apologize to those authors not cited here due to space limitations.
Mario García-Domínguez*Andalusian Centre for Molecular Biology and Regenerative Medicine-CABIMER CSIC-Universidad de Sevilla-Universidad Pablo de Olavide, Seville, Spain
*Correspondence to: Mario García-Domínguez,PhD, mario.garcia@cabimer.es.https://orcid.org/0000-0003-2211-8731(Mario García-Domínguez)
Date of submission: June 22, 2021
Date of decision: July 17, 2021
Date of acceptance: August 5, 2021
Date of web publication: January 7, 2022
https://doi.org/10.4103/1673-5374.332134
How to cite this article:García-Domínguez M(2022) Proper progression of neurogenesis relies on a defined pattern of SUMO-modified proteins.Neural Regen Res 17(8):1731-1732.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
?Article author(s) (unless otherwise stated in the text of the article) 2022. All rights reserved.No commercial use is permitted unless otherwise expressly granted.
- 中國神經(jīng)再生研究(英文版)的其它文章
- Motor neuron replacement therapy for amyotrophic lateral sclerosis
- Calorie restriction or dietary restriction: how far they can protect the brain against neurodegenerative diseases?
- Monodelphis domestica: a new source of mammalian primary neurons in vitro
- Vascular inflammation in the central nervous system
- Elucidating the pathological mechanisms of neurodegeneration in the lethal serpinopathy FENIB
- Glial cell line-derived neurotrophic factor in brain repair after focal ischemic stroke